1LOZ
 
 | |
1LST
 
 | |
22GS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y49F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A.J. | | Deposit date: | 1998-03-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamic description of the effect of the mutation Y49F on human glutathione transferase P1-1 in binding with glutathione and the inhibitor S-hexylglutathione.
J.Biol.Chem., 278, 2003
|
|
240D
 
 | |
1LR6
 
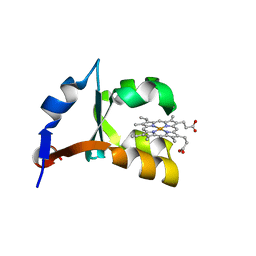 | | Crystal structure of V45Y mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Gan, J.-H, Wu, J, Wang, Z.-Q, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-05-15 | | Release date: | 2002-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of V45E and V45Y mutants and structure comparison of a variety of cytochrome b5 mutants.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2AE7
 
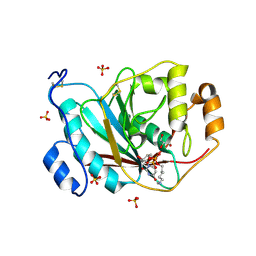 | | Crystal Structure of Human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4GAL-T1) in Complex with Pentasaccharide | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
1LQB
 
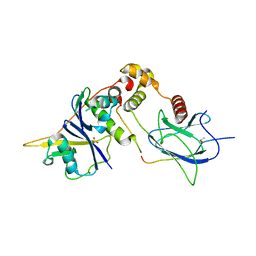 | | Crystal structure of a hydroxylated HIF-1 alpha peptide bound to the pVHL/elongin-C/elongin-B complex | | Descriptor: | Elongin B, Elongin C, Hypoxia-inducible factor 1 ALPHA, ... | | Authors: | Hon, W.C, Wilson, M.I, Harlos, K, Claridge, T.D, Schofield, C.J, Pugh, C.W, Maxwell, P.H, Ratcliffe, P.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2002-05-09 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL.
Nature, 417, 2002
|
|
1LSU
 
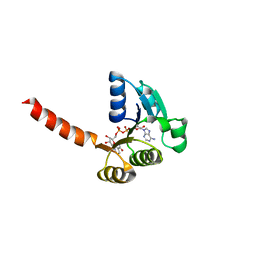 | | KTN Bsu222 Crystal Structure in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Conserved hypothetical protein yuaA | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
1LT6
 
 | |
1LUA
 
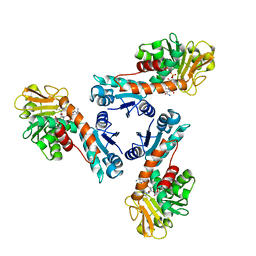 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 complexed with NADP | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
1LTO
 
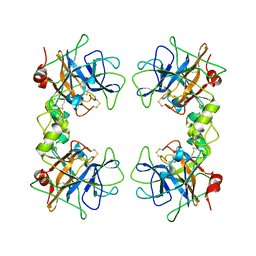 | | Human alpha1-tryptase | | Descriptor: | alpha tryptase I | | Authors: | Marquardt, U, Zettl, F, Huber, R, Bode, W, Sommerhoff, C.P. | | Deposit date: | 2002-05-20 | | Release date: | 2003-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of Human alpha1-Tryptase Reveals a Blocked Substrate-binding Region
J.MOL.BIOL., 321, 2002
|
|
1LMW
 
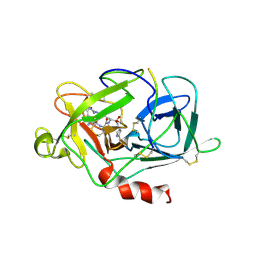 | | LMW U-PA Structure complexed with EGRCMK (GLU-GLY-ARG Chloromethyl Ketone) | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Spraggon, G.S, Phillips, C, Nowak, U.K, Ponting, C.P, Saunders, D, Dobson, C.M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1995-07-26 | | Release date: | 1996-01-29 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the catalytic domain of human urokinase-type plasminogen activator.
Structure, 3, 1995
|
|
2A0L
 
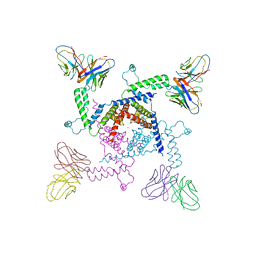 | | Crystal structure of KvAP-33H1 Fv complex | | Descriptor: | 33H1 Fv fragment, POTASSIUM ION, Voltage-gated potassium channel | | Authors: | Lee, S.Y, Lee, A, Chen, J, Mackinnon, R. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1LO0
 
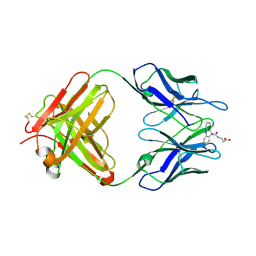 | | Catalytic Retro-Diels-Alderase Transition State Analogue Complex | | Descriptor: | 3-{[(9-CYANO-9,10-DIHYDRO-10-METHYLACRIDIN-9-YL)CARBONYL]AMINO}PROPANOIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-05 | | Release date: | 2002-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LPN
 
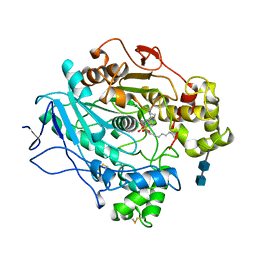 | |
2A2G
 
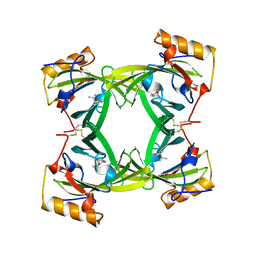 | | THE CRYSTAL STRUCTURES OF A2U-GLOBULIN AND ITS COMPLEX WITH A HYALINE DROPLET INDUCER. | | Descriptor: | D-LIMONENE 1,2-EPOXIDE, PROTEIN (ALPHA-2U-GLOBULIN) | | Authors: | Chaudhuri, B.N, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1998-11-19 | | Release date: | 1999-08-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structures of alpha 2u-globulin and its complex with a hyaline droplet inducer.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1LPM
 
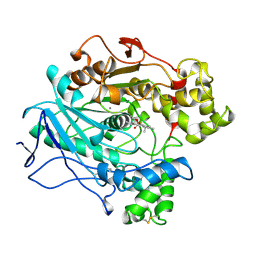 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-06 | | Release date: | 1995-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
2A2U
 
 | |
1LTT
 
 | |
1LUF
 
 | | Crystal Structure of the MuSK Tyrosine Kinase: Insights into Receptor Autoregulation | | Descriptor: | muscle-specific tyrosine kinase receptor musk | | Authors: | Till, J.H, Becerra, M, Watty, A, Lu, Y, Ma, Y, Neubert, T.A, Burden, S.J, Hubbard, S.R. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation.
Structure, 10, 2002
|
|
1LPH
 
 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-06-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
1LEO
 
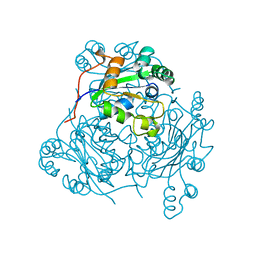 | | P100S NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Xu, Y, Veron, M. | | Deposit date: | 1996-05-22 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleoside diphosphate kinase. Investigation of the intersubunit contacts by site-directed mutagenesis and crystallography.
J.Biol.Chem., 271, 1996
|
|
2AD6
 
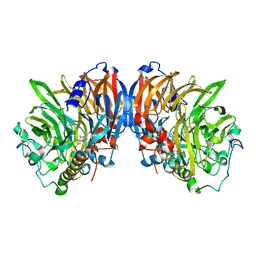 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
1LFO
 
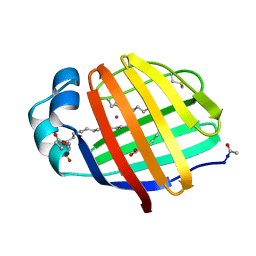 | | LIVER FATTY ACID BINDING PROTEIN-OLEATE COMPLEX | | Descriptor: | BUTENOIC ACID, LIVER FATTY ACID BINDING PROTEIN, OLEIC ACID, ... | | Authors: | Thompson, J, Winter, N, Terwey, D, Bratt, J, Banaszak, L. | | Deposit date: | 1996-12-09 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the liver fatty acid-binding protein. A complex with two bound oleates.
J.Biol.Chem., 272, 1997
|
|
1LNH
 
 | |
