7ADT
 
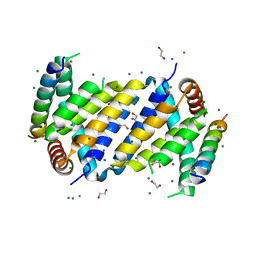 | | Orf virus Apoptosis inhibitor ORFV125 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis inhibitor, Apoptosis regulator BAX, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.210037 Å) | | Cite: | Crystal structures of ORFV125 provide insight into orf virus-mediated inhibition of apoptosis.
Biochem.J., 477, 2020
|
|
7A8Q
 
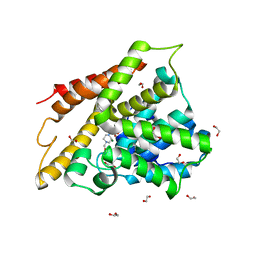 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-654 | | Descriptor: | 1,2-ETHANEDIOL, 2-cycloheptyl-5-[4-methoxy-3-[[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]methoxy]phenyl]-4,4-dimethyl-pyrazolidin-3-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-654
To be published
|
|
8CCH
 
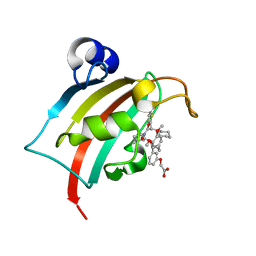 | | The Fk1 domain of FKBP51 in complex with 2-(3-((1R)-1-(((2S)-1-(2-cyclohexyl-2-(thiophen-2-yl)acetyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-thiophen-2-yl-ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
7A9V
 
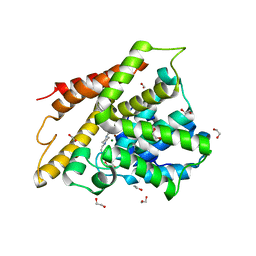 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-635 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-[4-methoxy-3-(2-phenylethoxy)phenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-02 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-635
To be published
|
|
7AB9
 
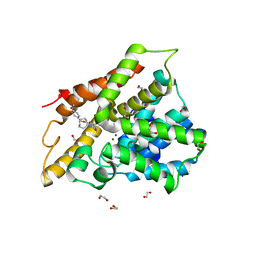 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-656 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-{4-methoxy-3-[2-(4-methoxyphenyl)ethoxy]phenyl}-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-656
To be published
|
|
3TZW
 
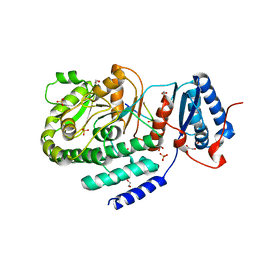 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in the orthorhombic apoform at 2.6 A | | Descriptor: | 1,2-ETHANEDIOL, 12-mer peptide, Polyketide synthase PKS13, ... | | Authors: | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | Deposit date: | 2011-09-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
8QPG
 
 | | Turret of Haloferax tailed virus 1 | | Descriptor: | MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ZINC ION, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
6SX7
 
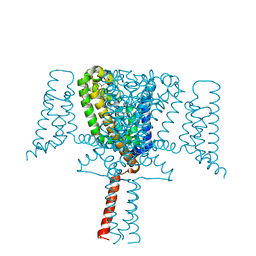 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) (2.2 Angstrom resolution) | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
7ABE
 
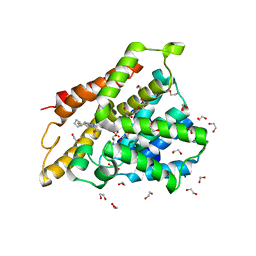 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-769 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-[3-(cyclopentyloxy)-4-methoxyphenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-769
To be published
|
|
4Y7F
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and 3-(phosphonooxy)propanoic acid (PPA) - GpgS Mn2+ UDP-Glc PPA | | Descriptor: | 1,2-ETHANEDIOL, 3-(phosphonooxy)propanoic acid, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.231 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3HCA
 
 | | Crystal Structure of E185Q hPNMT in Complex With Octopamine and AdoHcy | | Descriptor: | 1,2-ETHANEDIOL, 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
4YK2
 
 | |
4MSQ
 
 | | Crystal structure of Schizosaccharomyces pombe AMSH-like protease sst2 catalytic domain bound to ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease sst2, PHOSPHATE ION, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
4E9F
 
 | | Structure of the glycosylase domain of MBD4 bound to AP site containing DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*CP*GP*(3DR)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2012-03-21 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
7O2O
 
 | |
7A6I
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with LDC8201 | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[5-[4-chloranyl-2-[4-(4-methylpiperazin-1-yl)phenyl]-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]-2-methyl-phenyl]propanamide | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-25 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
7A6K
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with TAK-788 | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-25 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
3U6T
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, KANAMYCIN A, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A
To be Published
|
|
3U70
 
 | | Crystal structure of type 1 ribosome inactivating protein complexed with adenine in low ionic solvent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of type 1 ribosome inactivating protein complexed with adenine in low ionic solvent
TO BE PUBLISHED
|
|
4N4V
 
 | |
6ZX3
 
 | | OMPD-domain of human UMPS in complex with 6-thiocarboxamido-UMP at 1.15 Angstroms resolution | | Descriptor: | GLYCEROL, PROLINE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Schimdt, T. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
8QQN
 
 | |
8QE6
 
 | |
5FL4
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, ACETIC ACID, CARBONIC ANHYDRASE 9, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
8QSY
 
 | | Portal capsid interface of full Haloferax tailed virus 1. | | Descriptor: | Capsid stabilization protein, HK97 gp5-like major capsid protein, HK97 gp6-like/SPP1 gp15-like head-tail connector, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-11 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
