6VCO
 
 | | Crystal structure of E.coli RppH in complex with ppcpA | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, RNA pyrophosphohydrolase | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
5T2C
 
 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
8DF0
 
 | | Abp1D receptor binding domain | | Descriptor: | Abp1D Receptor Binding Domain | | Authors: | Tamadonfar, K.O, Pinkner, J.S, Dodson, K.W, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8IJ7
 
 | |
4ZXU
 
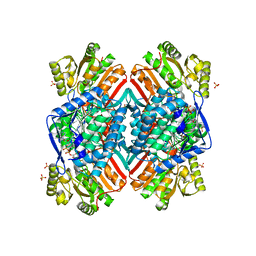 | | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289.
To be Published
|
|
8DKA
 
 | |
3B9L
 
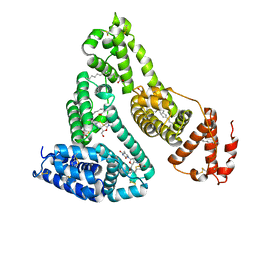 | | Human serum albumin complexed with myristate and AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, MYRISTIC ACID, Serum albumin | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
6VHE
 
 | |
8DEY
 
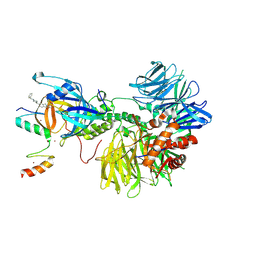 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
5T13
 
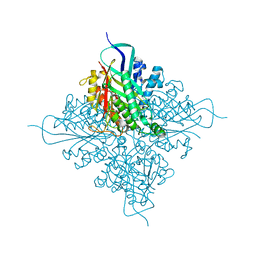 | |
8A9Q
 
 | | Computational design of stable mammalian serum albumins for bacterial expression | | Descriptor: | Albumin, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Khersonsky, O, Dym, O, Fleishman, J.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stable Mammalian Serum Albumins Designed for Bacterial Expression.
J.Mol.Biol., 435, 2023
|
|
8CWL
 
 | | Cryo-EM structure of Human 15-PGDH in complex with small molecule SW222746 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)], 2-methyl-6-[7-(piperidine-1-carbonyl)quinoxalin-2-yl]isoquinolin-1(2H)-one | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-05-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
8R37
 
 | | Klebsiella pneumoniae fosfomycin-resistance protein (FosAKP) | | Descriptor: | FOSFOMYCIN, FosA family fosfomycin resistance glutathione transferase, L(+)-TARTARIC ACID, ... | | Authors: | Papageorgiou, A.C, Varotsou, C, Labrou, N.E. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Studies of Klebsiella pneumoniae Fosfomycin-Resistance Protein and Its Application for the Development of an Optical Biosensor for Fosfomycin Determination.
Int J Mol Sci, 25, 2023
|
|
8D3Z
 
 | | Crystal structure of GalS1 in complex with Manganese from Populus trichocarpas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Galactan synthase, ... | | Authors: | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
8RNX
 
 | | Hen Egg White Lysozyme soaked with [HIsq][trans-RuCl4(DMSO)(Isq)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8D3T
 
 | | Crystal structure of GalS1 from Populus trichocarpas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
5AED
 
 | | A bacterial protein structure in glycoside hydrolase family 31 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-GLUCOSIDASE YIHQ, CALCIUM ION | | Authors: | Jin, Y, Speciale, G, Davies, G.J, Williams, S.J, Goddard-Borger, E.D. | | Deposit date: | 2015-08-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Yihq is a Sulfoquinovosidase that Cleaves Sulfoquinovosyl Diacylglyceride Sulfolipids.
Nat.Chem.Biol., 12, 2016
|
|
6VCB
 
 | | Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator | | Descriptor: | 1-[(1R)-1-(2,6-dichloro-3-methoxyphenyl)ethyl]-6-{2-[(2R)-piperidin-2-yl]phenyl}-1H-benzimidazole, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Feng, D, Bueno, A, Kobilka, B, Sloop, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into probe-dependent positive allosterism of the GLP-1 receptor.
Nat.Chem.Biol., 16, 2020
|
|
5T9G
 
 | |
5T2U
 
 | | short chain dehydrogenase/reductase family protein | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Mickael, B, Van Wyk, N, Baneres-Roquet, F, Yann, G, Laurent, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of NADP(+) triggers an open-to-closed transition in a mycobacterial FabG beta-ketoacyl-ACP reductase.
Biochem. J., 474, 2017
|
|
6VCL
 
 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
8DJD
 
 | |
1UOD
 
 | | Crystal structure of the dihydroxyacetone kinase from E. coli in complex with dihydroxyacetone-phosphate | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCERALDEHYDE-3-PHOSPHATE, SULFATE ION | | Authors: | Siebold, C, Garcia-Alles, L.F, Luthi-Nyffeler, T, Flukiger-Bruhwiler, K, Burgi, H.-B, Baumann, U, Erni, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-09-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoenolpyruvate- and ATP-Dependent Dihydroxyacetone Kinases: Covalent Substrate-Binding and Kinetic Mechanism
Biochemistry, 43, 2004
|
|
8DJE
 
 | | CRYSTAL STRUCTURE OF GLYCOGEN SYNTHASE KINASE 3 BETA COMPLEXED WITH 3-[(CYCLOPROPYLMETHYL)AMINO] -N-(4-PHENYLPYRIDIN-3-YL)IMIDAZO[1,2-B]PYRIDAZINE-8-CARBOX AMIDE | | Descriptor: | (4S)-3-[(cyclopropylmethyl)amino]-N-(4-phenylpyridin-3-yl)imidazo[1,2-b]pyridazine-8-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Muckelbauer, J.K. | | Deposit date: | 2022-06-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Design, Structure-Activity Relationships, and In Vivo Evaluation of Potent and Brain-Penetrant Imidazo[1,2- b ]pyridazines as Glycogen Synthase Kinase-3 beta (GSK-3 beta ) Inhibitors.
J.Med.Chem., 66, 2023
|
|
5TA7
 
 | | Crystal structure of BuGH117Bwt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
