3PSJ
 
 | |
2F9S
 
 | | 2nd Crystal Structure Of A Soluble Domain Of ResA In The Oxidised Form | | Descriptor: | Thiol-disulfide oxidoreductase resA | | Authors: | Colbert, C.L, Wu, Q, Erbel, P.J.A, Gardner, K.H, Deisenhofer, J. | | Deposit date: | 2005-12-06 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Mechanism of substrate specificity in Bacillus subtilis ResA, a thioredoxin-like protein involved in cytochrome c maturation
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7L0O
 
 | |
4NY3
 
 | | Human PTPA in complex with peptide | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein phosphatase 2A activator, ... | | Authors: | Loew, C, Quistgaard, E.M, Nordlund, P. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural basis for PTPA interaction with the invariant C-terminal tail of PP2A.
Biol.Chem., 395, 2014
|
|
439D
 
 | | 5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3' | | Descriptor: | BARIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3') | | Authors: | Perbandt, M, Lorenz, S, Vallazza, M, Erdmann, V.A, Betzel, C. | | Deposit date: | 1999-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2X8A
 
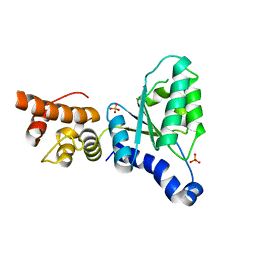 | | Human Nuclear Valosin containing protein Like (NVL), C-terminal AAA- ATPase domain | | Descriptor: | NUCLEAR VALOSIN-CONTAINING PROTEIN-LIKE, PHOSPHATE ION | | Authors: | Moche, M, Schuetz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Kraulis, P, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, VanDenBerg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H. | | Deposit date: | 2010-03-08 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Nuclear Valosin Containing Protein Like (Nvl) , C-Terminal Aaa-ATPase Domain
To be Published
|
|
3MMO
 
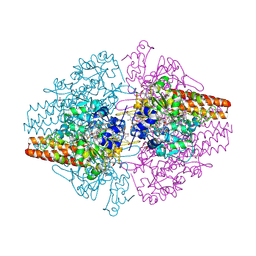 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with cyanide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-04-20 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of complexes of octahaem cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens with sulfite and cyanide.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1CSE
 
 | |
5SFM
 
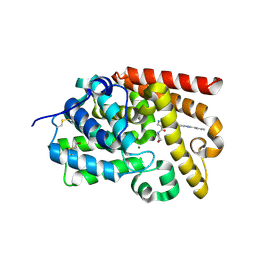 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n3cn1c(nc(n1)c2ccccc2)cc3NC(=O)c5c(C(=O)N4CCOCC4)cnn5C, micromolar IC50=0.0012349 | | Descriptor: | 1-methyl-4-(morpholine-4-carbonyl)-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-c]pyrimidin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Lerner, C, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5ITJ
 
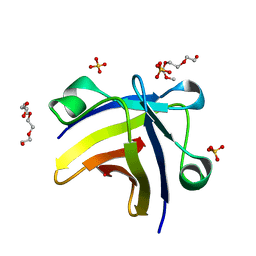 | | The structure of histone-like protein | | Descriptor: | AbrB family transcriptional regulator, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
6I18
 
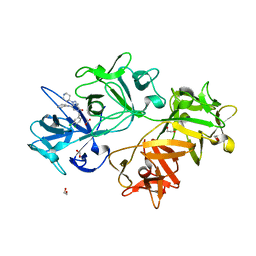 | | CRYSTAL STRUCTURE OF FASCIN IN COMPLEX WITH BDP-13176 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3,4-dichlorophenyl)methyl]-4-oxidanylidene-1-piperidin-4-yl-~{N}-pyridin-4-yl-pyrazolo[4,3-c]pyridine-7-carboxamide, ACETATE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2018-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-based design, synthesis and biological evaluation of a novel series of isoquinolone and pyrazolo[4,3-c]pyridine inhibitors of fascin 1 as potential anti-metastatic agents.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2VUZ
 
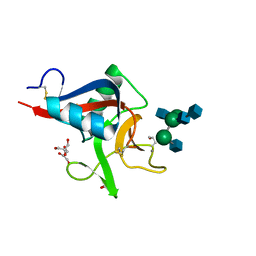 | | Crystal structure of Codakine in complex with biantennary nonasaccharide at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
5ITM
 
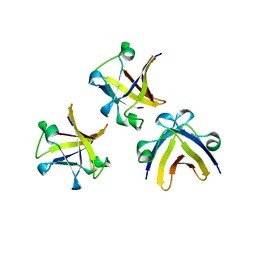 | | The structure of truncated histone-like protein | | Descriptor: | AbrB family transcriptional regulator | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
1Z60
 
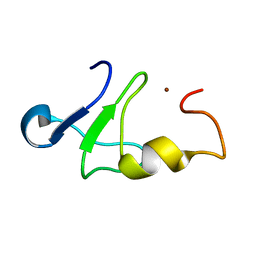 | | Solution structure of the carboxy-terminal domain of human TFIIH P44 subunit | | Descriptor: | TFIIH basal transcription factor complex p44 subunit, ZINC ION | | Authors: | Kellenberger, E, Dominguez, C, Fribourg, S, Wasielewski, E, Moras, D, Poterszman, A, Boelens, R, Kieffer, B. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions.
J.Biol.Chem., 280, 2005
|
|
4FZR
 
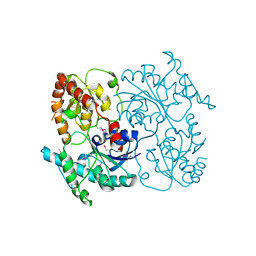 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4CDA
 
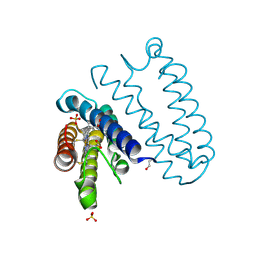 | | Spectroscopically-validated structure of ferric cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BJO
 
 | |
3IW4
 
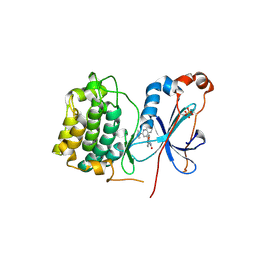 | | Crystal structure of PKC alpha in complex with NVP-AEB071 | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]-1H-pyrrole-2,5-dione, Protein kinase C alpha type | | Authors: | Stark, W, Rummel, G, Strauss, A, Cowan-Jacob, S.W. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes
J.Med.Chem., 52, 2009
|
|
4JMY
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with DDIVPC peptide | | Descriptor: | HCV non-structural protein 4A, Polyprotein, SODIUM ION, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Importance of the peptide scaffold of drugs that target the hepatitis C virus NS3 protease and its crucial bioactive conformation and dynamic factors.
To be Published
|
|
3Q9V
 
 | |
1UPL
 
 | | Crystal structure of MO25 alpha | | Descriptor: | MO25 PROTEIN | | Authors: | Milburn, C.C, Boudeau, J, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mo25 Alpha in Complex with the C-Terminus of the Pseudo Kinase Ste-20 Related Adaptor (Strad)
Nat.Struct.Mol.Biol., 11, 2004
|
|
1CXA
 
 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C, IMIDAZOLE | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-02-14 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
4CDY
 
 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 160K | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8AAG
 
 | | H1-bound palindromic nucleosome, state 1 | | Descriptor: | DNA/RNA (185-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Alegrio Louro, J, Beinsteiner, B, Cheng, T.C, Patel, A.K.M, Boopathi, R, Angelov, D, Hamiche, A, Bednar, J, Kale, S, Dimitrov, S, Klaholz, B. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Nucleosome dyad determines the H1 C-terminus collapse on distinct DNA arms.
Structure, 31, 2023
|
|
