4M4M
 
 | | The structure of Ni T6 bovine insulin | | Descriptor: | Insulin, NICKEL (II) ION | | Authors: | Frankaer, C.G, Harris, P, Stahl, K. | | Deposit date: | 2013-08-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards accurate structural characterization of metal centres in protein crystals: the structures of Ni and Cu T6 bovine insulin derivatives.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1LVA
 
 | |
7AD1
 
 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(One up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein,Spike glycoprotein,Envelope glycoprotein,SARS-CoV-2 S protein | | Authors: | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
9F43
 
 | |
4X3H
 
 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4MD2
 
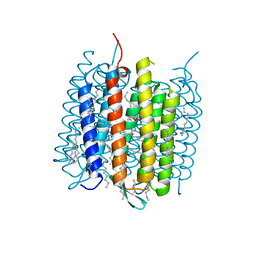 | | Ground state of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9BAF
 
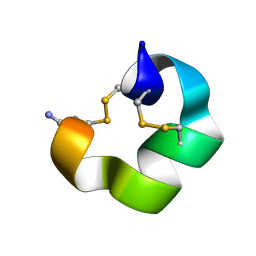 | | Solution NMR structure of conofurin-Delta | | Descriptor: | Alpha-conotoxin LvIA | | Authors: | Harvey, P.J, Craik, D.J, Hone, A.J, McIntosh, J.M. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Novel Peptide Derivatives of the Severe Acute Respiratory Syndrome-Coronavirus-2 Spike-Protein that Potently Inhibit Nicotinic Acetylcholine Receptors.
J.Med.Chem., 67, 2024
|
|
9BB4
 
 | |
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
9F45
 
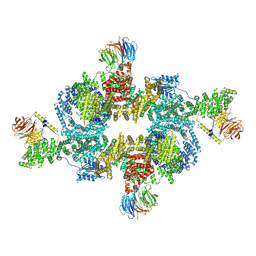 | | cryo-EM structure of human LST2 bound to human mTOR complex 1 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Lateral signaling target protein 2 homolog, Regulatory-associated protein of mTOR, ... | | Authors: | Craigie, L.M, Maier, T. | | Deposit date: | 2024-04-26 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | mTORC1 phosphorylates and stabilizes LST2 to negatively regulate EGFR.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9CIH
 
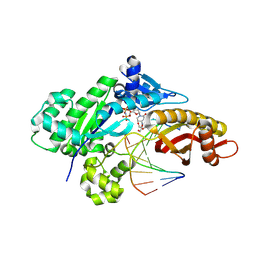 | |
1LWJ
 
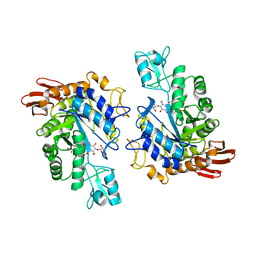 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE/ACARBOSE COMPLEX | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, MODIFIED ACARBOSE PENTASACCHARIDE | | Authors: | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | Deposit date: | 2002-05-31 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2FFF
 
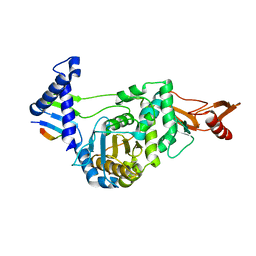 | |
2QO5
 
 | | Crystal structure of the cysteine 91 threonine mutant of zebrafish liver bile acid-binding protein complexed with cholic acid | | Descriptor: | CHOLIC ACID, Liver-basic fatty acid binding protein | | Authors: | Capaldi, S, Saccomani, G, Perduca, M, Monaco, H.L. | | Deposit date: | 2007-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Single Amino Acid Mutation in Zebrafish (Danio rerio) Liver Bile Acid-binding Protein Can Change the Stoichiometry of Ligand Binding.
J.Biol.Chem., 282, 2007
|
|
9B9G
 
 | | Structure of the PI4KA complex bound to Calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, Hyccin, ... | | Authors: | Shaw, A.L, Suresh, S, Yip, C.K, Burke, J.E. | | Deposit date: | 2024-04-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of calcineurin bound to PI4KA reveals dual interface in both PI4KA and FAM126A.
Structure, 2024
|
|
6OYA
 
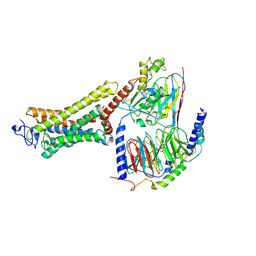 | | Structure of the Rhodopsin-Transducin-Nanobody Complex | | Descriptor: | Camelid antibody VHH fragment, Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
1LYY
 
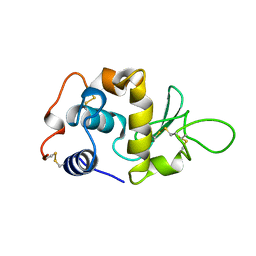 | |
9BB3
 
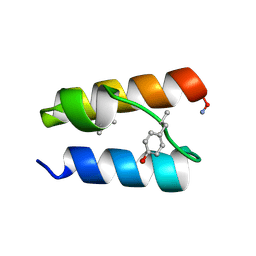 | |
5WXE
 
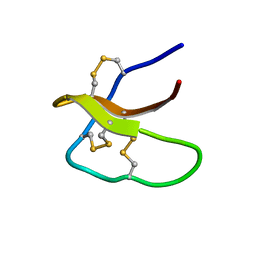 | | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers | | Descriptor: | jasmintide js3 | | Authors: | Kumari, G, Wong, K.H, Serra, A, Shin, J, Yoon, H.S, Sze, S.K, James, P.T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers
To Be Published
|
|
1M63
 
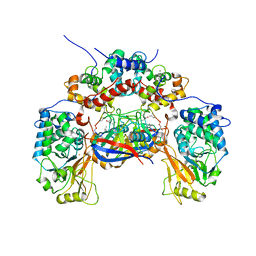 | | Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes | | Descriptor: | CALCINEURIN B SUBUNIT ISOFORM 1, CALCIUM ION, CYCLOSPORIN A, ... | | Authors: | Huai, Q, Kim, H.-Y, Liu, Y, Zhao, Y, Mondragon, A, Liu, J.O, Ke, H. | | Deposit date: | 2002-07-12 | | Release date: | 2002-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Calcineurin-Cyclophilin-Cyclosporin Shows Common But Distinct Recognition of Immunophilin-Drug Complexes
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LZO
 
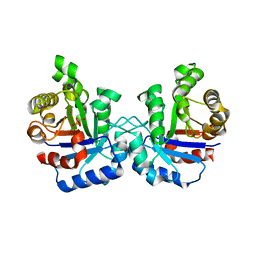 | | Plasmodium Falciparum Triosephosphate Isomerase-Phosphoglycolate Complex | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate Isomerase | | Authors: | Parthasarathy, S, Balaram, H, Balaram, P, Murthy, M.R. | | Deposit date: | 2002-06-11 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Plasmodium falciparum triosephosphate isomerase-phosphoglycolate complex in two crystal forms: characterization of catalytic loop open and closed conformations in the ligand-bound state
Biochemistry, 41, 2002
|
|
5WQZ
 
 | |
6OD5
 
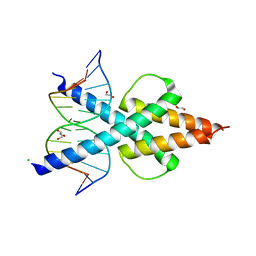 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
9BB1
 
 | |
