2CEN
 
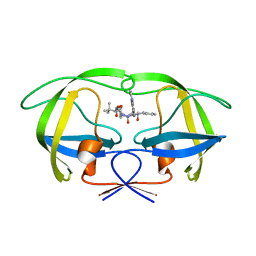 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
4YDL
 
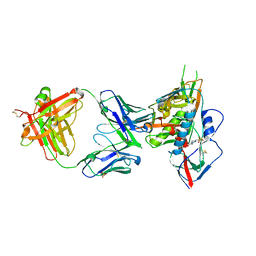 | | Crystal structure of broadly and potently neutralizing antibody C38-VRC18.02 in complex with HIV-1 clade AE strain 93TH057gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
5AHA
 
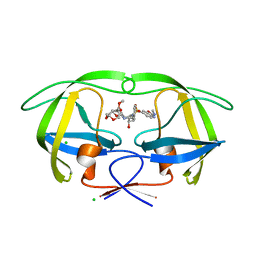 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5DO6
 
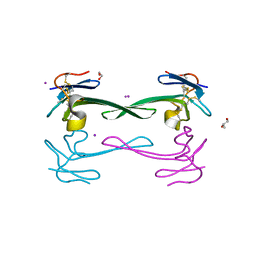 | | Crystal structure of Dendroaspis polylepis venom mambalgin-1 T23A mutant | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Mambalgin-1, ... | | Authors: | Stura, E.A, Tepshi, L, Kessler, P, Gilles, M, Servent, D. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
5ZGW
 
 | |
4BF1
 
 | | Three dimensional structure of human carbonic anhydrase II in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, CARBONIC ANHYDRASE 2, SODIUM ION, ... | | Authors: | Tars, K, Leitans, J, Zalubovskis, R. | | Deposit date: | 2013-03-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 5-Substituted-(1,2,3-Triazol-4-Yl)Thiophene-2-Sulfonamides Strongly Inhibit Human Carbonic Anhydrases I, II, Ix and Xii: Solution and X-Ray Crystallographic Studies.
Bioorg.Med.Chem., 21, 2013
|
|
5AH8
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(3,3,3-trifluoropropoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AHB
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH6
 
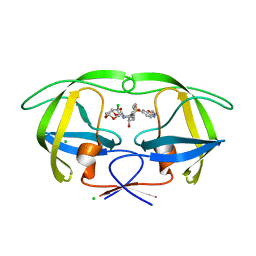 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH7
 
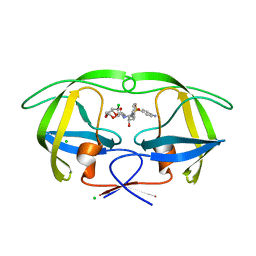 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5B4O
 
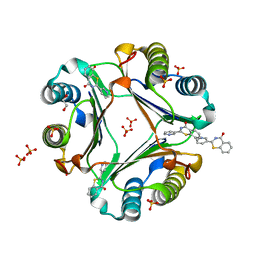 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
1ZAF
 
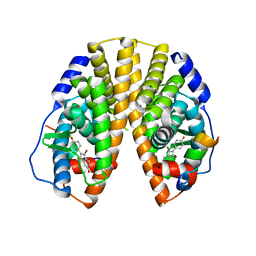 | | Crystal structure of estrogen receptor beta complexed with 3-Bromo-6-hydroxy-2-(4-hydroxy-phenyl)-inden-1-one | | Descriptor: | 3-BROMO-6-HYDROXY-2-(4-HYDROXYPHENYL)-1H-INDEN-1-ONE, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | McDevitt, R.E, Malamas, M.S, Manas, E.S, Unwalla, R.J, Xu, Z.B, Miller, C.P, Harris, H.A. | | Deposit date: | 2005-04-06 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Estrogen receptor ligands: design and synthesis of new 2-arylindene-1-ones
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2XIY
 
 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2A4F
 
 | | Synthesis and Activity of N-Axyl Azacyclic Urea HIV-1 Protease Inhibitors with High Potency Against Multiple Drug Resistant Viral Strains. | | Descriptor: | (5R,6R)-5-BENZYL-6-HYDROXY-2,4-BIS(4-HYDROXY-3-METHOXYBENZYL)-1-[3-(4-HYDROXYPHENYL)PROPANOYL]-1,2,4-TRIAZEPAN-3-ONE, Pol polyprotein | | Authors: | Zhao, C, Sham, H, Sun, M, Lin, S, Stoll, V, Stewart, K.D, Mo, H, Vasavanonda, S, Saldivar, A, McDonald, E. | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and activity of N-acyl azacyclic urea HIV-1 protease inhibitors with high potency against multiple drug resistant viral strains
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6K9U
 
 | | Discovery of Pyrazolo[1,5-a]pyrimidine Derivative as a Highly Selective PDE10A Inhibitor | | Descriptor: | 2-(3,7-dimethylquinoxalin-2-yl)-~{N}-(oxan-4-yl)-5-pyrrolidin-1-yl-pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Takedomi, K, Koizumi, Y. | | Deposit date: | 2019-06-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a pyrazolo[1,5-a]pyrimidine derivative (MT-3014) as a highly selective PDE10A inhibitor via core structure transformation from the stilbene moiety.
Bioorg.Med.Chem., 27, 2019
|
|
5ZZV
 
 | | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution
To Be Published
|
|
4AWE
 
 | | The Crystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goncalves, A.M.D, Silva, C.S, De Sanctis, D, Bento, I. | | Deposit date: | 2012-06-01 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Endo-Beta-D-1,4-Mannanase from Chrysonilia Sitophila Displays a Novel Loop Arrangement for Substrate Selectivity
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2VS4
 
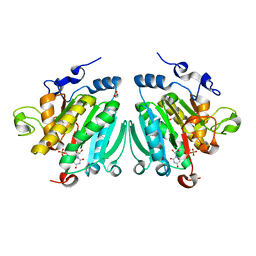 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
6KZC
 
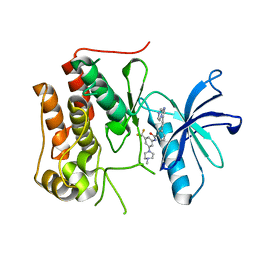 | | crystal structure of TRKc in complex with 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-N-(3-((4- methylpiperazin-1-yl)methyl)-5- (trifluoromethyl)phenyl)benzamide | | Descriptor: | 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]benzamide, NT-3 growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
3I7B
 
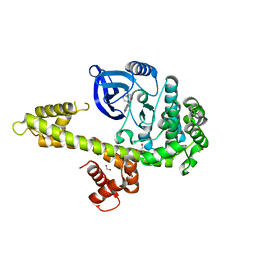 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with bumped kinase inhibitor NM-PP1 | | Descriptor: | 1,2-ETHANEDIOL, 1-(1-methylethyl)-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2009-07-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1ZY9
 
 | |
4Q6R
 
 | |
4QAG
 
 | | Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNASE H domain of HIV-1 reverse transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Himmel, D.M, Ho, W.C, Arnold, E. | | Deposit date: | 2014-05-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs.
J.Mol.Biol., 426, 2014
|
|
2VXM
 
 | | Screening a Limited Structure-based Library Identifies UDP-GalNAc- Specific Mutants of alpha-1,3 Galactosyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Acharya, K.R, Brew, K. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Screening a Limited Structure-Based Library Identifies Udp-Galnac-Specific Mutants of {Alpha}-1,3-Galactosyltransferase.
Glycobiology, 18, 2008
|
|
2WBB
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-BROMO-1,3-THIAZOL-2(3H)-YLIDENE]CARBAMOYL}-4-METHYLBENZENESULFONAMIDE | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
