5QUA
 
 | | Crystal Structure of swapped human Nck SH3.1 domain, 1.5A, C2221 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU1
 
 | | Crystal Structure of the monomeric human Nck SH3.1 domain, triclinic, 1.08A | | Descriptor: | Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU5
 
 | | Domain Swap in the first SH3 domain of human Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU2
 
 | | Crystal Structure of human Nck SH3.1 in complex with peptide PPPVPNPDY | | Descriptor: | ACE-PRO-PRO-PRO-VAL-PRO-ASN-PRO-ASP-TYR-NH2, Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU7
 
 | |
5QU4
 
 | |
1OOT
 
 | |
1OV3
 
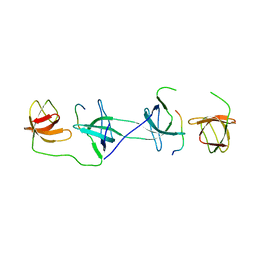 | | Structure of the p22phox-p47phox complex | | Descriptor: | Flavocytochrome b558 alpha polypeptide, Neutrophil cytosol factor 1 | | Authors: | Groemping, Y, Lapouge, K, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2003-03-25 | | Release date: | 2003-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of phosphorylation-induced activation of the NADPH oxidase
Cell(Cambridge,Mass.), 113, 2003
|
|
1NYF
 
 | |
1NLP
 
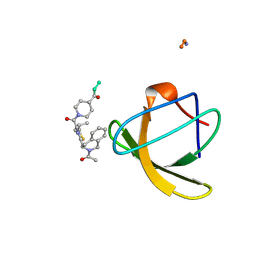 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-SRC, NL2 (MN8-MN1-PLPPLP) | | Authors: | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | Deposit date: | 1996-08-04 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
1NLO
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-SRC, NL1 (MN7-MN2-MN1-PLPPLP) | | Authors: | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | Deposit date: | 1996-08-04 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
1NYG
 
 | |
1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
4HXJ
 
 | | Crystal structure of SH3:RGT complex | | Descriptor: | C-terminal 3-mer peptide from Integrin beta-3, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Xiao, R, Meng, G. | | Deposit date: | 2012-11-11 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural framework of c-Src activation by integrin beta 3
Blood, 121, 2013
|
|
1NM7
 
 | | Solution structure of the ScPex13p SH3 domain | | Descriptor: | Peroxisomal Membrane Protein PAS20 | | Authors: | Pires, J.R, Hong, X, Brockmann, C, Volkmer-Engert, R, Schneider-Mergener, J, Oschkinat, H, Erdmann, R. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The ScPex13p SH3 Domain Exposes Two Distinct Binding Sites for Pex5p and Pex14p
J.Mol.Biol., 326, 2003
|
|
1OEB
 
 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
4GLM
 
 | | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens] | | Descriptor: | Dynamin-binding protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Guan, X, Huang, H, Tempel, W, Gu, J, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens]
to be published
|
|
4GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1QKW
 
 | | Alpha-spectrin Src Homology 3 domain, N47G mutant in the distal loop. | | Descriptor: | ALPHA II SPECTRIN, GLYCEROL, SULFATE ION | | Authors: | Vega, M.C, Martinez, J, Serrano, L. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural characterization of Asn and Ala residues in the disallowed II' region of the Ramachandran plot.
Protein Sci., 9, 2000
|
|
4J9E
 
 | |
1QLY
 
 | | NMR Study of the SH3 Domain From Bruton's Tyrosine Kinase, 20 Structures | | Descriptor: | TYROSINE-PROTEIN KINASE BTK | | Authors: | Tzeng, S.R, Lou, Y.C, Pai, M.T, Chen, C, Chen, S.H, Cheng, J.Y. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Btk SH3 Domain Complexed with a Proline-Rich Peptide from P120Cbl
J.Biomol.NMR, 16, 2000
|
|
4J9B
 
 | |
4J9D
 
 | |
4JJB
 
 | |
