1QBY
 
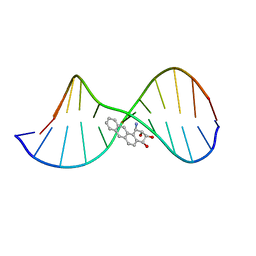 | | THE SOLUTION STRUCTURE OF A BAY-REGION 1R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*GP*GP*AP*CP*(BZA)AP*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP* G)-3' | | Authors: | Li, Z, Mao, H, Kim, H.-Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-04-27 | | Release date: | 1999-05-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (-)-(1R,2S,3R, 4S)-N6-[1-benz[a]anthracenyl]-2'-deoxyadenosyl adduct in an oligodeoxynucleotide containing the human N-ras codon 61 sequence.
Biochemistry, 38, 1999
|
|
8I4X
 
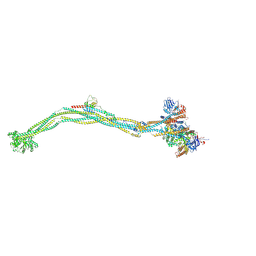 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3L15
 
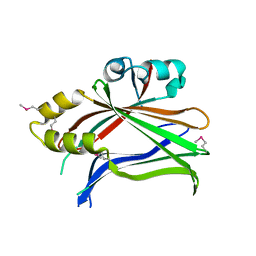 | | Human Tead2 transcriptional factor | | Descriptor: | GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Tomchick, D.R, Luo, X, Tian, W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the YAP-binding domain of human TEAD2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4JL9
 
 | | Crystal structure of mouse TBK1 bound to BX795 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Li, P, Shu, C. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.0999 Å) | | Cite: | Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity.
Structure, 21, 2013
|
|
1VY4
 
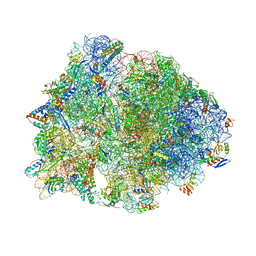 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing acylated tRNA-substrates in the A and P sites. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3DU6
 
 | |
3DB3
 
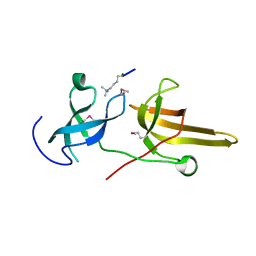 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
1LX7
 
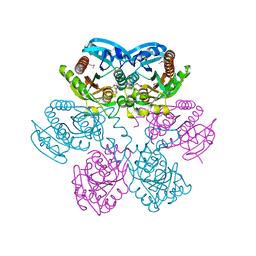 | | Structure of E. coli uridine phosphorylase at 2.0A | | Descriptor: | uridine phosphorylase | | Authors: | Burling, T, Buglino, J.A, Kniewel, R, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-04 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli uridine phosphorylase at 2.0 A.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8C3N
 
 | |
4JLC
 
 | | Crystal structure of mouse TBK1 bound to SU6668 | | Descriptor: | 3-{2,4-dimethyl-5-[(Z)-(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-1H-pyrrol-3-yl}propanoic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Li, P. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity.
Structure, 21, 2013
|
|
7BCY
 
 | |
7BED
 
 | |
1PVO
 
 | |
1UB4
 
 | |
3GCB
 
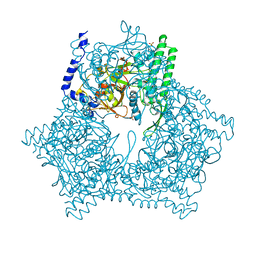 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A/DELTAK454 | | Descriptor: | GAL6, GLYCEROL, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
3DU5
 
 | |
2AXZ
 
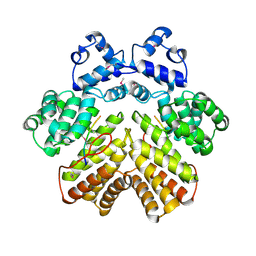 | | Crystal structure of PrgX/cCF10 complex | | Descriptor: | LVTLVFV peptide, PrgX, TPPKEVT(MSE) peptide | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AXV
 
 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1A77
 
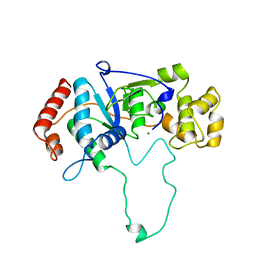 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1BHL
 
 | | CACODYLATED CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BI4
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-22 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
3EUL
 
 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1XXC
 
 | |
4HCZ
 
 | | PHF1 Tudor in complex with H3K36me3 | | Descriptor: | H3L-like histone, PHD finger protein 1 | | Authors: | Musselman, C.A, Roy, S, Nunez, J, Kutateladze, T.G. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
