2QX7
 
 | |
6UE0
 
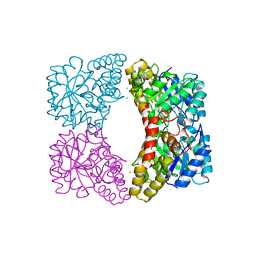 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
6I38
 
 | |
6I4H
 
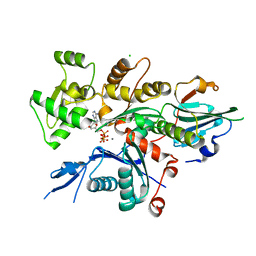 | | Crystal Structure of Plasmodium falciparum actin I (F54Y mutant) in the Ca-ATP state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CALCIUM ION, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6ULU
 
 | | D104A/S128A S. typhimurium siroheme synthase | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-10-08 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
6UMX
 
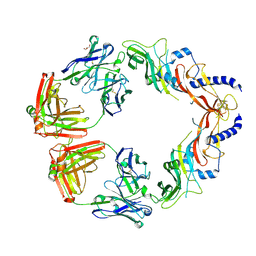 | | Structural basis for specific inhibition of extracellular activation of pro/latent myostatin by SRK-015 | | Descriptor: | GL29H4-16 Fab Heavy Chain,GL29H4-16 Fab Heavy Chain, GL29H4-16 Fab Light Chain,GL29H4-16 Fab Light Chain, GLYCEROL, ... | | Authors: | Dagbay, K.B, Treece, E, Streich Jr, F.C, Jackson, J.W, Faucette, R.R, Nikiforov, A, Lin, S.C, Bostion, C.J, Nicholls, S.B, Capili, A.D, Carven, G.J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of specific inhibition of extracellular activation of pro- or latent myostatin by the monoclonal antibody SRK-015.
J.Biol.Chem., 295, 2020
|
|
6TX0
 
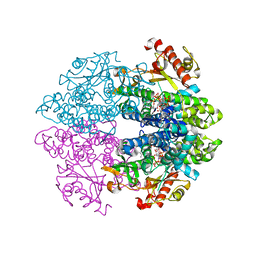 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TV4
 
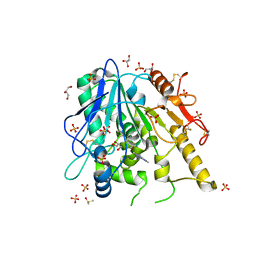 | | CFF-Notum complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Caffeine inhibits Notum activity by binding at the catalytic pocket.
Commun Biol, 3, 2020
|
|
8BWK
 
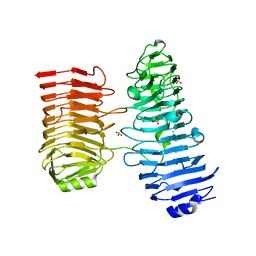 | |
6ZWX
 
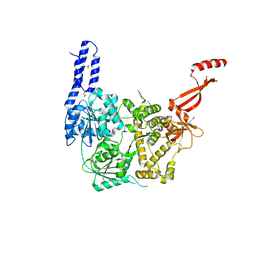 | | Crystal structure of E. coli RNA helicase HrpA | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8BX1
 
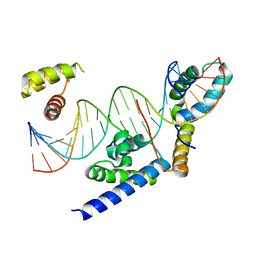 | |
2C0N
 
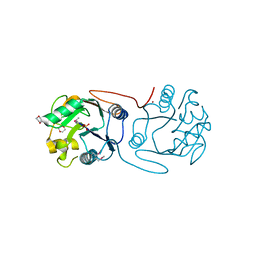 | | Crystal Structure of A197 from STIV | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, A197, NICKEL (II) ION, ... | | Authors: | Larson, E.T, Reiter, D, Young, M, Lawrence, C.M. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-29 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of A197 from Sulfolobus Turreted Icosahedral Virus: A Crenarchaeal Viral Glycosyltransferase Exhibiting the Gt-A Fold.
J.Virol., 80, 2006
|
|
5R4H
 
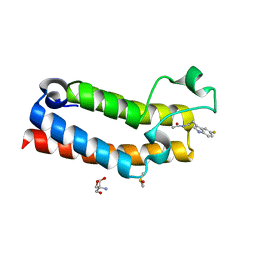 | | PanDDA analysis group deposition -- CRYSTAL STRUCTURE OF THE BROMODOMAIN OF HUMAN NUCLEOSOME-REMODELING FACTOR SUBUNIT BPTF in complex with FMOPL000287a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, Nucleosome-remodeling factor subunit BPTF, ... | | Authors: | Talon, R, Krojer, T, Fairhead, M, Sethi, R, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Wright, N, MacLean, E, Renjie, Z, Dias, A, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2020-02-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2YPW
 
 | | Atomic model for the N-terminus of TraO fitted in the full-length structure of the bacterial pKM101 type IV secretion system core complex | | Descriptor: | TRAO | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-02 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.4 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
3V99
 
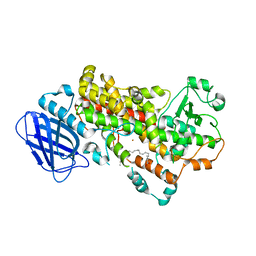 | | S663D Stable-5-LOX in complex with Arachidonic Acid | | Descriptor: | ARACHIDONIC ACID, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Gilbert, N.C, Rui, Z, Neau, D.B, Waight, M, Bartlett, S.G, Boeglin, W.E, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2011-12-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Conversion of human 5-lipoxygenase to a 15-lipoxygenase by a point mutation to mimic phosphorylation at Serine-663.
Faseb J., 26, 2012
|
|
3VBH
 
 | | Crystal structure of formaldehyde treated human enterovirus 71 (space group R32) | | Descriptor: | CHLORIDE ION, Genome Polyprotein, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2BQ4
 
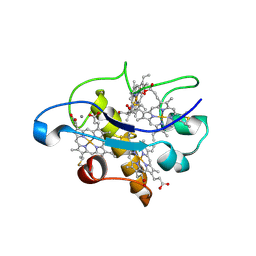 | | Crystal structure of type I cytochrome c3 from Desulfovibrio africanus | | Descriptor: | BASIC CYTOCHROME C3, CALCIUM ION, HEME C | | Authors: | Czjzek, M, Pieulle, L, Morelli, X, Guerlesquin, F, Hatchikian, E.C. | | Deposit date: | 2005-04-27 | | Release date: | 2005-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Type I / Type II Cytochrome C(3) Complex: An Electron Transfer Link in the Hydrogen-Sulfate Reduction Pathway.
J.Mol.Biol., 354, 2005
|
|
6E0A
 
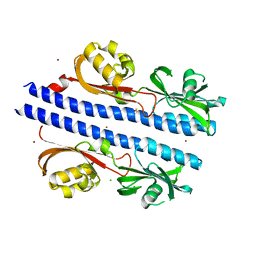 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-07-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
8BBJ
 
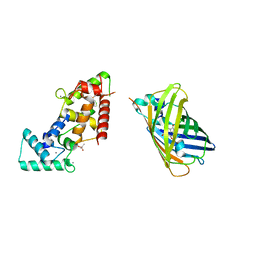 | |
8BAN
 
 | | Secretagogin (mouse) in complex with its target peptide from SNAP-25 | | Descriptor: | CALCIUM ION, Green fluorescent protein,Synaptosomal-associated protein 25, Secretagogin | | Authors: | Schnell, R, Szodorai, E. | | Deposit date: | 2022-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A hydrophobic groove in secretagogin allows for alternate interactions with SNAP-25 and syntaxin-4 in endocrine tissues.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6E0G
 
 | | Mitochondrial peroxiredoxin from Leishmania infantum after heat stress without unfolding client protein | | Descriptor: | mitochondrial 2-cys-peroxiredoxin | | Authors: | Teixeira, F, Tse, E, Makepeace, K.A.T, Borchers, C.H, Castro, H, Tomas, A.M, Poole, L.B, Southworth, D.R, Jakob, U. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Chaperone activation and client binding of a 2-cysteine peroxiredoxin.
Nat Commun, 10, 2019
|
|
8BAV
 
 | | Secretagogin (human) in complex with its target peptide from SNAP-25 | | Descriptor: | ACETATE ION, CALCIUM ION, Green fluorescent protein,Synaptosomal-associated protein 25, ... | | Authors: | Schnell, R, Szodorai, E. | | Deposit date: | 2022-10-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A hydrophobic groove in secretagogin allows for alternate interactions with SNAP-25 and syntaxin-4 in endocrine tissues.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8BWH
 
 | |
8BWE
 
 | |
6OAF
 
 | | Sudan virus nucleoprotein core domain in complex with VP35 chaperoning peptide | | Descriptor: | Polymerase cofactor VP35,Nucleoprotein | | Authors: | Landeras-Bueno, S, Oda, S, Norris, M.J, Li Salie, Z, Ollmann Saphire, E. | | Deposit date: | 2019-03-15 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sudan Ebolavirus VP35-NP Crystal Structure Reveals a Potential Target for Pan-Filovirus Treatment.
Mbio, 10, 2019
|
|
