2PP8
 
 | | Formate bound to oxidized wild type AfNiR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (I) ION, ... | | Authors: | Tocheva, E.I, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2007-04-28 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved active site residues limit inhibition of a copper-containing nitrite reductase by small molecules.
Biochemistry, 47, 2008
|
|
5WYJ
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
2Y0I
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH TANKYRASE-2 (TNKS2) FRAGMENT PEPTIDE (21-MER) | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-12-02 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains.
FEBS J., 278, 2011
|
|
1BPW
 
 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ALDEHYDE DEHYDROGENASE) | | Authors: | Johansson, K, El Ahmad, M, Ramaswamy, S, Hjelmqvist, L, Jornvall, H, Eklund, H. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
3SFE
 
 | | crystal structure of porcine mitochondrial respiratory complex II bound with oxaloacetate and thiabendazole | | Descriptor: | 2-(1,3-THIAZOL-4-YL)-1H-BENZIMIDAZOLE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Zhou, Q.J, Zhai, Y.J, Liu, M, Sun, F. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature.
Protein Cell, 2, 2011
|
|
4GUG
 
 | | 1.62 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant in Complex with Dehydroshikimate (Crystal Form #1) | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
4GVX
 
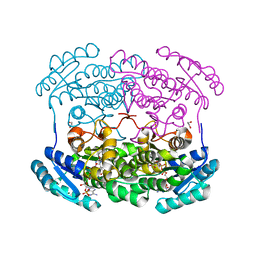 | | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NADP and L-fucose | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Hobbs, M.E, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NADP and L-fucose
To be Published
|
|
2FNW
 
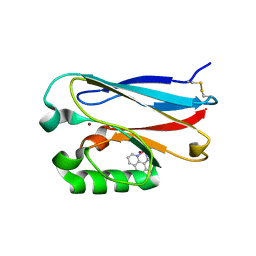 | | Pseudomonas aeruginosa E2Q/H83Q/M109H-azurin RE(PHEN)(CO)3 | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-01-11 | | Release date: | 2006-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Excited-state dynamics of structurally characterized [ReI(CO)3(phen)(HisX)]+ (X = 83, 109) Pseudomonas aeruginosa azurins in aqueous solution.
J.Am.Chem.Soc., 128, 2006
|
|
1OAO
 
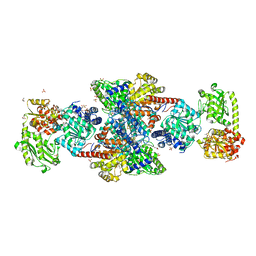 | | NiZn[Fe4S4] and NiNi[Fe4S4] clusters in closed and open alpha subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase | | Descriptor: | ACETATE ION, BICARBONATE ION, CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Darnault, C, Volbeda, A, Kim, E.J, Legrand, P, Vernede, X, Lindahl, P.A, Fontecilla-Camps, J.C. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-11 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] Clusters in Closed and Open Alpha Subunits of Acetyl-Coa Synthase/Carbon Monoxide Dehydrogenase
Nat.Struct.Biol., 10, 2003
|
|
5SAR
 
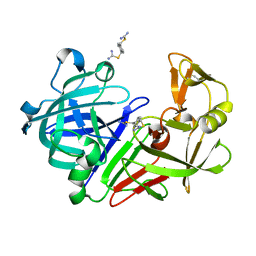 | | Endothiapepsin in complex with compound FU290-1 | | Descriptor: | (1,4-phenylene)bis(methylene) dicarbamimidothioate, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3LG4
 
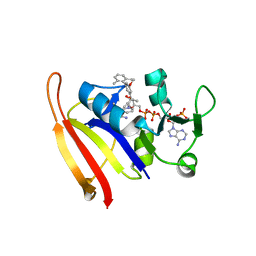 | | Staphylococcus aureus V31Y, F92I mutant dihydrofolate reductase complexed with NADPH and 5-[(3S)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine | | Descriptor: | 5-[(3S)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Frey, K.M, Anderson, A.C. | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Predicting resistance mutations using protein design algorithms.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5XDX
 
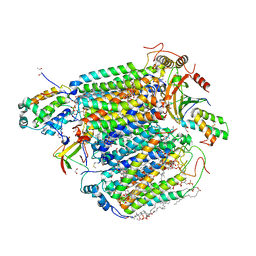 | | Bovine heart cytochrome c oxidase in the reduced state with pH 7.3 at 1.99 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-03-30 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of bovine cytochrome c oxidase in the ligand-free reduced state at neutral pH.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4H97
 
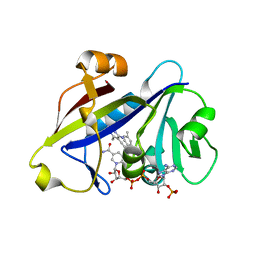 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5-(4-methylphenyl)phenyl]but-1-yn-1-yl}-6-methylpyrimidine-2,4-diamine (UCP111D4M) | | Descriptor: | 5-[(3R)-3-(5-methoxy-4'-methylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate Reductase, GLYCEROL, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-09-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1C4W
 
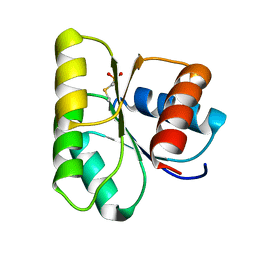 | | 1.9 A STRUCTURE OF A-THIOPHOSPHONATE MODIFIED CHEY D57C | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Halkides, C.J, McEvoy, M.M, Matsumura, P, Volz, K, Dahlquist, F.W. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY.
Biochemistry, 39, 2000
|
|
7KXA
 
 | |
1GCO
 
 | | CRYSTAL STRUCTURE OF GLUCOSE DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | GLUCOSE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glucose dehydrogenase from Bacillus megaterium IWG3 at 1.7 A resolution.
J.Biochem., 129, 2001
|
|
5UP7
 
 | | Crystal Structure of the Ni-bound Human Heavy-Chain Ferritin 122H-delta C-star variant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
4GL4
 
 | | Crystal structure of oxidized S-nitrosoglutathione reductase from Arabidopsis thalina, complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase class-3, SULFATE ION, ... | | Authors: | Weichsel, A, Crotty, J, Montfort, W.R. | | Deposit date: | 2012-08-13 | | Release date: | 2013-02-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and kinetic behavior of alcohol dehydrogenase III/S-nitrosoglutathione reductase from arabidopsis thalina
To be Published
|
|
1FX2
 
 | |
4GUJ
 
 | | 1.50 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase, ZINC ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
5VU0
 
 | | Crystal structure of the complex between afucosylated/galactosylated human IgG1 Fc and Fc gamma receptor IIIa (CD16A) with Man5 N-glycans | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Subedi, G.S, Marcella, A.M, Barb, A.W. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Antibody Fucosylation Lowers the Fc gamma RIIIa/CD16a Affinity by Limiting the Conformations Sampled by the N162-Glycan.
ACS Chem. Biol., 13, 2018
|
|
1M1N
 
 | | Nitrogenase MoFe protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(7)-MO-S(9)-N CLUSTER, ... | | Authors: | Einsle, O, Tezcan, F.A, Andrade, S.L.A, Schmid, B, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-19 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor.
Science, 297, 2002
|
|
5S9S
 
 | |
4I4H
 
 | | Crystal structure of CYP3A4 ligated to pyridine-substituted desoxyritonavir | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, pyridin-3-ylmethyl [(2R,5S)-5-{[N-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-D-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pyridine-Substituted Desoxyritonavir Is a More Potent Inhibitor of Cytochrome P450 3A4 than Ritonavir.
J.Med.Chem., 56, 2013
|
|
4HPI
 
 | | Crystal Structure of Hen Egg White Lysozyme complex with GN2-M | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-1-DEOXYNOJIRIMYCIN, Lysozyme C | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-10-23 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A novel transition-state analogue for lysozyme, 4-O-beta-tri-N-acetylchitotriosyl moranoline, provided evidence supporting the covalent glycosyl-enzyme intermediate.
J.Biol.Chem., 288, 2013
|
|
