6ZOK
 
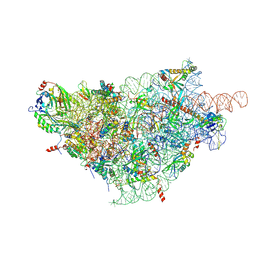 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6H02
 
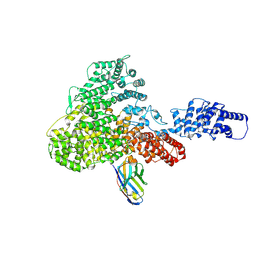 | |
1SMY
 
 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
5T2T
 
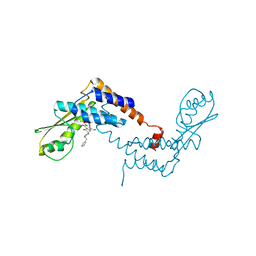 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease bound to compound L742001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-08-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
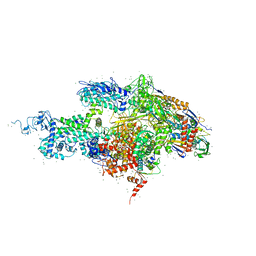
6ZOL
 
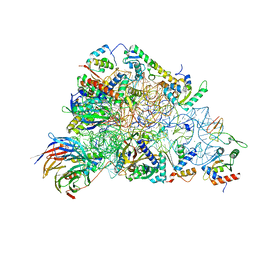 | | SARS-CoV-2-Nsp1-40S complex, focused on head | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7AAV
 
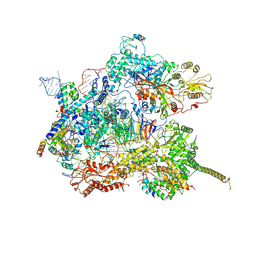 | | Human pre-Bact-2 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, D-chiro inositol hexakisphosphate, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7OMG
 
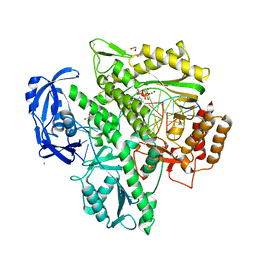 | | Crystal structure of KOD DNA Polymerase in a ternary complex with an Uracil containing template | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OMB
 
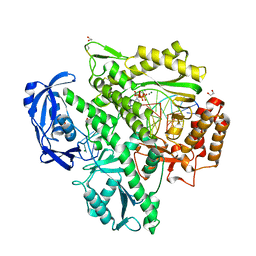 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
4JK1
 
 | |
4JK2
 
 | |
4G7Z
 
 | |
8D6J
 
 | |
8KG6
 
 | | Yeast replisome in state I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
7EH1
 
 | | Thermus thermophilus transcription initiation complex containing a template-strand purine at position TSS-2, GpG RNA primer, and CMPcPP | | Descriptor: | 1,4-BUTANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (27-MER), ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EH2
 
 | |
5UBP
 
 | | TREX2 M-region | | Descriptor: | 26S proteasome complex subunit SEM1, Leucine permease transcriptional regulator, Nuclear mRNA export protein THP1 | | Authors: | Stewart, M, Gordon, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Sac3 RNA-binding M-region in the Saccharomyces cerevisiae TREX-2 complex.
Nucleic Acids Res., 45, 2017
|
|
8KG9
 
 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
4XJN
 
 | | Structure of the parainfluenza virus 5 nucleocapsid-RNA complex: an insight into paramyxovirus polymerase activity | | Descriptor: | LEAD (II) ION, Nucleocapsid, RNA (78-MER) | | Authors: | Alayyoubi, M, Leser, G.P, Kors, C.A, Lamb, R.A. | | Deposit date: | 2015-01-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8E86
 
 | | Human DNA polymerase eta-DNA-rC-ended primer-dGMPNPP ternary mismatch complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8G
 
 | |
8E85
 
 | | Human DNA polymerase eta-DNA-rG-ended primer-dGMPNPP ternary mismatch complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*CP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8F
 
 | |
8E8E
 
 | |
8E8D
 
 | |
8E8C
 
 | |
