8G9S
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC8, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9T
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC9, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9U
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | CRISPR-associated protein, Csd1 family, Csd2 family, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAF
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAM
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GIS
 
 | |
8GIR
 
 | |
8GIO
 
 | |
8GIT
 
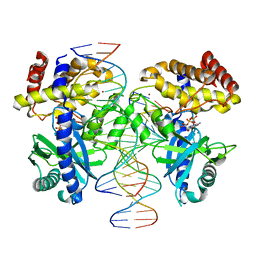 | |
8GIM
 
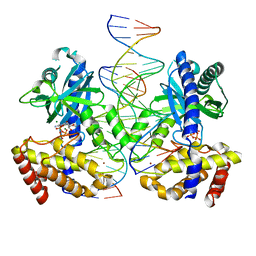 | |
8G1J
 
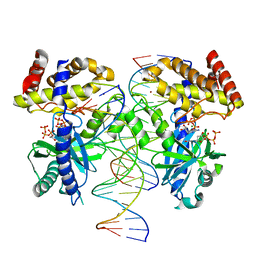 | |
8G10
 
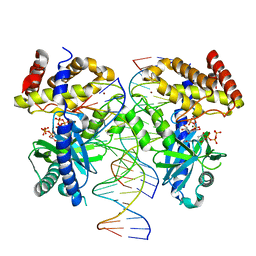 | |
8GIP
 
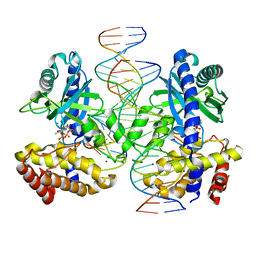 | |
1VDC
 
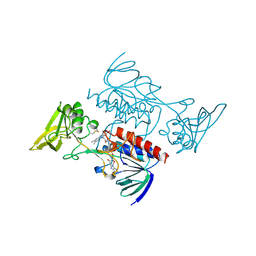 | | STRUCTURE OF NADPH DEPENDENT THIOREDOXIN REDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DEPENDENT THIOREDOXIN REDUCTASE, SULFATE ION | | Authors: | Dai, S, Eklund, H. | | Deposit date: | 1996-09-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Arabidopsis thaliana NADPH dependent thioredoxin reductase at 2.5 A resolution.
J.Mol.Biol., 264, 1996
|
|
8I1U
 
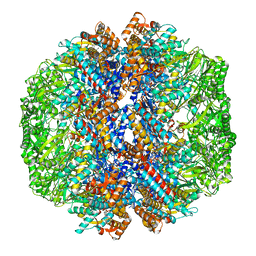 | | Human TRiC-PhLP2A complex in the closed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8HKW
 
 | |
4YXQ
 
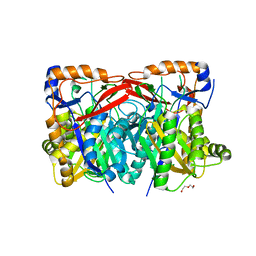 | |
1Q7S
 
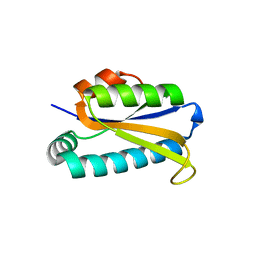 | | Crystal structure of bit1 | | Descriptor: | bit1 | | Authors: | De Pereda, J.M, Waas, W.F, Jan, Y, Ruoslahti, E, Schimmel, P, Pascual, J. | | Deposit date: | 2003-08-19 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity.
J.Biol.Chem., 279, 2004
|
|
2PIJ
 
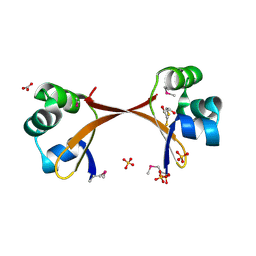 | | Structure of the Cro protein from prophage Pfl 6 in Pseudomonas fluorescens Pf-5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, Prophage Pfl 6 Cro, ... | | Authors: | Roessler, C.G, Roberts, S.A, Montfort, W.R, Cordes, M.H.J. | | Deposit date: | 2007-04-13 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4R8P
 
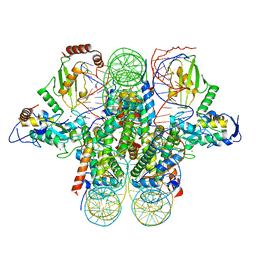 | | Crystal structure of the Ring1B/Bmi1/UbcH5c PRC1 ubiquitylation module bound to the nucleosome core particle | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase RING2, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | McGinty, R.K, Henrici, R.C, Tan, S. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2846 Å) | | Cite: | Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome.
Nature, 514, 2014
|
|
1TER
 
 | |
2APS
 
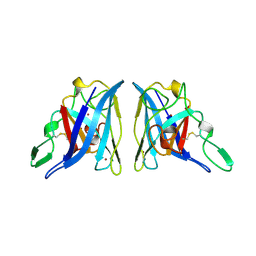 | | CU/ZN SUPEROXIDE DISMUTASE FROM ACTINOBACILLUS PLEUROPNEUMONIAE | | Descriptor: | COPPER (II) ION, PROTEIN (CU,ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Forest, K.T, Langford, P.R, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cu,Zn superoxide dismutase structure from a microbial pathogen establishes a class with a conserved dimer interface.
J.Mol.Biol., 296, 2000
|
|
8P5R
 
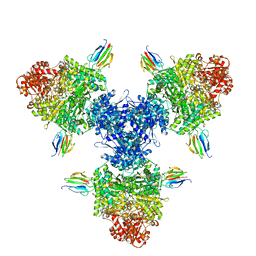 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
1B77
 
 | |
1UX6
 
 | |
