8DF5
 
 | | SARS-CoV-2 Beta RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McCallum, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution.
Science, 377, 2022
|
|
1YTN
 
 | | HYDROLASE | | Descriptor: | NITRATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Yuvaniyama, C, Fauman, E.B, Saper, M.A. | | Deposit date: | 1996-05-01 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structures of Yersinia tyrosine phosphatase with bound tungstate and nitrate. Mechanistic implications.
J.Biol.Chem., 271, 1996
|
|
8A5W
 
 | | Crystal structure of the human phosphoserine aminotransferase (PSAT) in complex with O-phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (2~{S})-2-[[(~{R})-[[(5~{S})-5-azanyl-6-oxidanylidene-hexyl]amino]-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methyl]amino]-3-phosphonooxy-propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | Deposit date: | 2022-06-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
8A5V
 
 | | Crystal structure of the human phosposerine aminotransferase (PSAT) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase, SULFATE ION | | Authors: | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | Deposit date: | 2022-06-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
7ZL9
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, FiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZLY
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
7ZI4
 
 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
7TKM
 
 | |
2JOC
 
 | | Mouse Itch 3rd domain phosphorylated in T30 | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
8DD2
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus Zolpidem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Hibbs, R.E. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and dynamic mechanisms of GABA A receptor modulators with opposing activities.
Nat Commun, 13, 2022
|
|
2JO9
 
 | | Mouse Itch 3rd WW domain complex with the Epstein-Barr virus latent membrane protein 2A derived peptide EEPPPPYED | | Descriptor: | Itchy E3 ubiquitin protein ligase, Latent membrane protein 2 | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
8DD3
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Hibbs, R.E. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and dynamic mechanisms of GABA A receptor modulators with opposing activities.
Nat Commun, 13, 2022
|
|
7TKP
 
 | |
7TKH
 
 | |
7TZ2
 
 | | Structure of human Fibrinogen-like protein 1 | | Descriptor: | CALCIUM ION, Fibrinogen-like protein 1 | | Authors: | Ming, Q, Tran, T.H, Luca, V.C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | LAG3 ectodomain structure reveals functional interfaces for ligand and antibody recognition.
Nat.Immunol., 23, 2022
|
|
7ZKA
 
 | | ABCB1 V978C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Urbatsch, I, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7TKR
 
 | |
7TKI
 
 | |
7ZK6
 
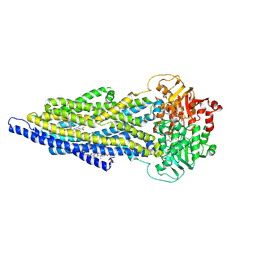 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to 2 molecules of AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7TKK
 
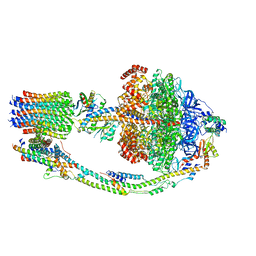 | |
7ZK5
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7TKG
 
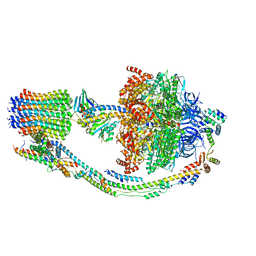 | |
7ZK9
 
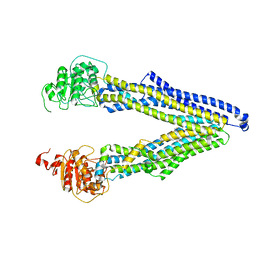 | | ABCB1 L971C mutant (mABCB1) in the inward facing state | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7TKQ
 
 | |
