3P5R
 
 | | Crystal Structure of Taxadiene Synthase from Pacific Yew (Taxus brevifolia) in complex with Mg2+ and 2-fluorogeranylgeranyl diphosphate | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, Taxadiene synthase | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis.
Nature, 469, 2011
|
|
5QTW
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-14-[(methoxycarbonyl)amino]-2,3,4,5,6,7-hexahydro-1H-8,11-epimino-1,9-benzodiazacyclotridecine-2-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5QTV
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-2-(trifluoromethyl)-2,3,4,5,6,7-hexahydro-1H-8,11-epimino-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5QTX
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR ethyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-14-[(methoxycarbonyl)amino]-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecine-2-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5Q0G
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-3-ethyl-2-oxo-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5Q0D
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-2-oxo-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7U63
 
 | | Crystal Structure Anti-Oxycodone Antibody HY2-A12 Fab Complexed with Oxycodone | | Descriptor: | 14-hydroxy-3-methoxy-17-methyl-5beta-4,5-epoxymorphinan-6-one, HY2-A12 Fab Heavy Chain, HY2-A12 Fab Light Chain | | Authors: | Rodarte, J.V, Pancera, M.P, Weidle, C, Rupert, P.B, Strong, R.K. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
5M35
 
 | |
1VJ6
 
 | |
1VFC
 
 | | Solution Structure Of The DNA Complex Of Human Trf2 | | Descriptor: | Short C-rich starnd, Short G-rich strand, Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
1VF9
 
 | | Solution Structure Of Human Trf2 | | Descriptor: | Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
1W1F
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-06-17 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Lyn-SH3 Domain in Complex with a Herpesviral Protein Reveals an Extended Recognition Motif that Enhances Binding Affinity.
Protein Sci., 14, 2005
|
|
5WKR
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference complexed with naringenin (ancCC) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Engineered Chalcone Isomerase ancCC, ... | | Authors: | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
5WL4
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancR3) | | Descriptor: | Engineered Chalcone Isomerase ancR3, FORMIC ACID | | Authors: | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
5WMM
 
 | | Crystal structure of an adenylation domain interrupted by a methylation domain (AMA4) from nonribosomal peptide synthetase TioS | | Descriptor: | (2S)-2-amino-3-methylbutanoyl (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl hydrogen (S)-phosphate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pang, A.H, Mori, S, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-07-30 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for backbone N-methylation by an interrupted adenylation domain.
Nat. Chem. Biol., 14, 2018
|
|
5WLP
 
 | |
5WYZ
 
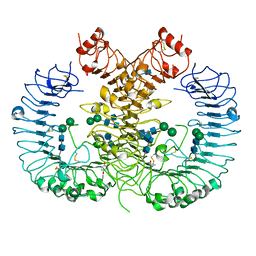 | | Crystal structure of human TLR8 in complex with CU-CPT9b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-methyl-4-oxidanyl-phenyl)quinolin-7-ol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
5WA9
 
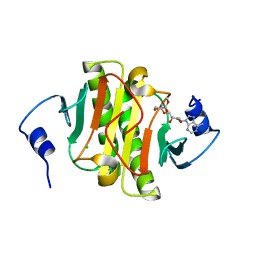 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Ala phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~{R})-1-methoxy-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5WEZ
 
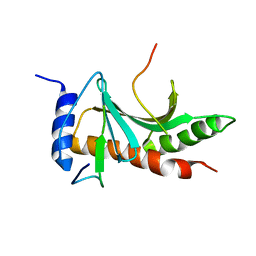 | | Structure of the Tir-CesT effector-chaperone complex | | Descriptor: | Tir chaperone, Translocated intimin receptor Tir | | Authors: | Little, D.J, Coombes, B.K. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular basis for CesT recognition of type III secretion effectors in enteropathogenic Escherichia coli.
PLoS Pathog., 14, 2018
|
|
5WPR
 
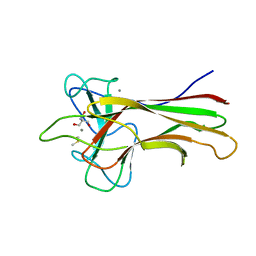 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5W96
 
 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | Descriptor: | Fz7 binding peptide | | Authors: | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | Deposit date: | 2017-06-22 | | Release date: | 2018-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
5WJP
 
 | |
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
3C4U
 
 | |
3C56
 
 | | Class II fructose-1,6-bisphosphate aldolase from helicobacter pylori in complex with N-(3-Hydroxypropyl)-glycolohydroxamic acid bisphosphate, a competitive inhibitor | | Descriptor: | 3-{hydroxy[(phosphonooxy)acetyl]amino}propyl dihydrogen phosphate, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Coincon, M, Sygusch, J, Potvin-Dulude, P. | | Deposit date: | 2008-01-30 | | Release date: | 2008-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Biochemical Evaluation of Selective Inhibitors of Class II Fructose Bisphosphate Aldolases: Towards New Synthetic Antibiotics.
Chemistry, 14, 2008
|
|
