7XQB
 
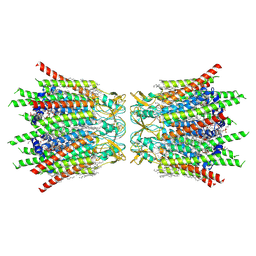 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQF
 
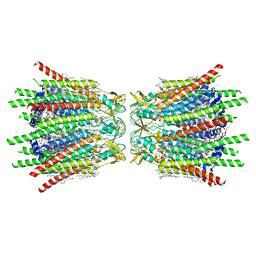 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQD
 
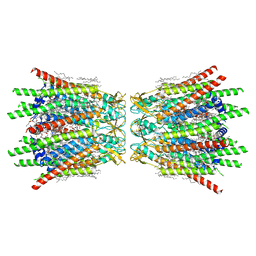 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7Y3F
 
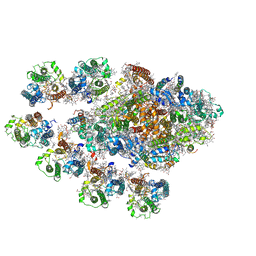 | | Structure of the Anabaena PSI-monomer-IsiA supercomplex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Nagao, R, Kato, K, Hamaguchi, T, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2022-06-10 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nat Commun, 14, 2023
|
|
7Y8A
 
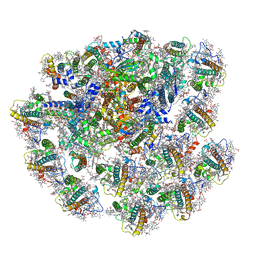 | | Cryo-EM structure of cryptophyte photosystem I | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Zhao, L.S, Zhang, Y.Z, Liu, L.N, Li, K. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis and evolution of the photosystem I-light-harvesting supercomplex of cryptophyte algae.
Plant Cell, 35, 2023
|
|
7Y7B
 
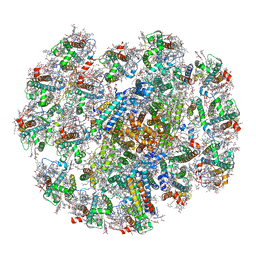 | | Cryo-EM structure of cryptophyte photosystem I | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Zhao, L.S, Li, K, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis and evolution of the photosystem I-light-harvesting supercomplex of cryptophyte algae.
Plant Cell, 35, 2023
|
|
7XQP
 
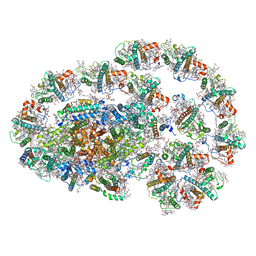 | | PSI-LHCI-LHCII-Lhcb9 supercomplex of Physcomitrella patens | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhang, S, Tang, K.L, Li, X.Y, Wang, W.D, Yan, Q.J, Shen, L.L, Kuang, T.Y, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
7YCA
 
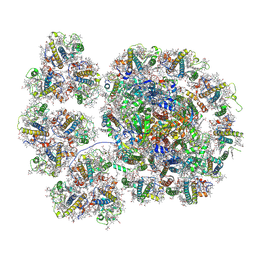 | | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
7Y41
 
 | |
4V9O
 
 | |
4U4N
 
 | | Crystal structure of Edeine bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4V44
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
4V50
 
 | | Crystal Structure of Ribosome with messenger RNA and the Anticodon stem-loop of P-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Berk, V, Zhang, W, Pai, R.D, Cate, J.H.D. | | Deposit date: | 2006-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for mRNA and tRNA positioning on the ribosome.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4U3M
 
 | | Crystal structure of Anisomycin bound to the yeast 80S ribosome | | Descriptor: | 18S rRNA, 25s rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4V64
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with hygromycin B. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2008-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for hygromycin B inhibition of protein biosynthesis
Rna, 14, 2008
|
|
4V9S
 
 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V8I
 
 | | Crystal structure of YfiA bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-12 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
4V98
 
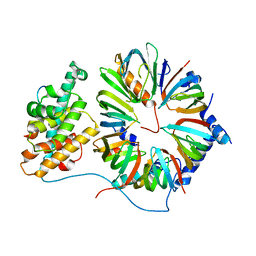 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
4UDF
 
 | | STRUCTURAL BASIS OF HUMAN PARECHOVIRUS NEUTRALIZATION BY HUMAN MONOCLONAL ANTIBODIES | | Descriptor: | HUMAN MONOCLONAL ANTIBODY, Protein VP0, Protein VP3 | | Authors: | Shakeel, S, Westerhuis, B.M, Ora, A, Koen, G, Bakker, A.Q, Claassen, Y, Beaumont, T, Wolthers, K.C, Butcher, S.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis of Human Parechovirus Neutralization by Human Monoclonal Antibodies.
J. Virol., 89, 2015
|
|
4U26
 
 | | Crystal structure of the E. coli ribosome bound to dalfopristin and quinupristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4V7S
 
 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V19
 
 | | Structure of the large subunit of the mammalian mitoribosome, part 1 of 2 | | Descriptor: | MAGNESIUM ION, MITORIBOSOMAL 16S RRNA, MITORIBOSOMAL CP TRNA, ... | | Authors: | Greber, B.J, Boehringer, D, Leibundgut, M, Bieri, P, Leitner, A, Schmitz, N, Aebersold, R, Ban, N. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Complete Structure of the Large Subunit of the Mammalian Mitochondrial Ribosome
Nature, 515, 2014
|
|
4V8A
 
 | | The structure of thermorubin in complex with the 70S ribosome from Thermus thermophilus. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D, Johnson, F.A, Steitz, T.A. | | Deposit date: | 2011-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge b2a of the ribosome.
J.Mol.Biol., 416, 2012
|
|
4V8R
 
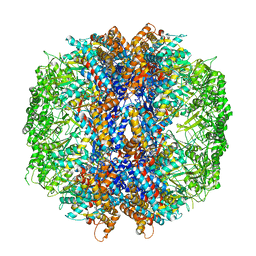 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Kalisman, N, Schroder, G.F, Levitt, M. | | Deposit date: | 2012-03-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
