3E8V
 
 | | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis | | Descriptor: | Possible transglutaminase-family protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis
To be Published
|
|
2D1F
 
 | | Structure of Mycobacterium tuberculosis threonine synthase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine synthase | | Authors: | Covarrubias, A.S, Bergfors, T, Mannerstedt, K, Oscarson, S, Jones, T.A, Mowbray, S.L, Hogbom, M. | | Deposit date: | 2005-08-20 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, biochemical, and in vivo investigations of the threonine synthase from Mycobacterium tuberculosis.
J.Mol.Biol., 381, 2008
|
|
3ED4
 
 | | Crystal structure of putative arylsulfatase from escherichia coli | | Descriptor: | ARYLSULFATASE, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Gilmore, M, Chang, S, Bain, K, Wasserman, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arylsulfatase from Escherichia Coli
To be Published
|
|
3EER
 
 | | High resolution structure of putative organic hydroperoxide resistance protein from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, IMIDAZOLE, Organic hydroperoxide resistance protein, ... | | Authors: | Nocek, B, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-05 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the organic hydroperoxide
resistance protein from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
5T44
 
 | | Crystal structure of Frizzled 7 CRD | | Descriptor: | Frizzled-7 | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannous, R.H, Wang, W. | | Deposit date: | 2016-08-29 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.9944 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5SWI
 
 | | Crystal structure of SpGH92 in complex with mannose | | Descriptor: | CALCIUM ION, GLYCEROL, Sugar hydrolase, ... | | Authors: | Shapiro-Ward, S, Boraston, A.B. | | Deposit date: | 2016-08-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular Characterization of N-glycan Degradation and Transport in Streptococcus pneumoniae and Its Contribution to Virulence.
PLoS Pathog., 13, 2017
|
|
5SVH
 
 | |
5SWB
 
 | |
2CZJ
 
 | | Crystal structure of the tRNA domain of tmRNA from Thermus thermophilus HB8 | | Descriptor: | SsrA-binding protein, tmRNA (63-MER) | | Authors: | Bessho, Y, Shibata, R, Sekine, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3C4W
 
 | |
3BV9
 
 | | Structure of Thrombin Bound to the Inhibitor FM19 | | Descriptor: | FM19 inhibitor, GLYCEROL, IODIDE ION, ... | | Authors: | Nieman, M.T, Burke, F, Warnock, M, Zhou, Y, Sweigert, J, Chen, A, Ricketts, D, Lucchesi, B.R, Chen, Z, Di Cera, E, Hilfinger, J, Mosberg, H.I, Schmaier, A.H. | | Deposit date: | 2008-01-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thrombostatin FM compounds: direct thrombin inhibitors - mechanism of action in vitro and in vivo.
J.Thromb.Haemost., 6, 2008
|
|
5CTA
 
 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 10% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Esterase | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
3C4Z
 
 | |
3BOG
 
 | | Snow Flea Antifreeze Protein Quasi-racemate | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein, UNKNOWN LIGAND | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
1I7X
 
 | | BETA-CATENIN/E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, EPITHELIAL-CADHERIN | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
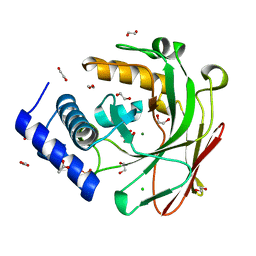
5D2H
 
 | |
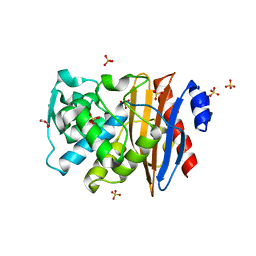
1O7E
 
 | |
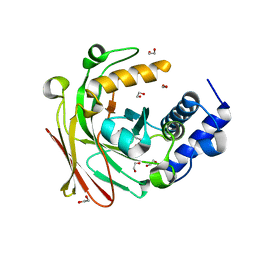
5D2K
 
 | |
2C9E
 
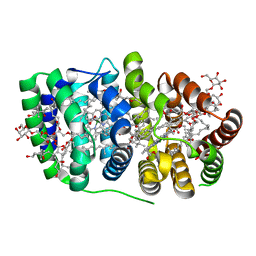 | | Peridinin-chlorophyll a protein, high-salt form | | Descriptor: | CHLOROPHYLL A, DIGALACTOSYL DIACYL GLYCEROL (DGDG), MAGNESIUM ION, ... | | Authors: | Schulte, T, Sharples, F.P, Hiller, R.G, Hofmann, E. | | Deposit date: | 2005-12-09 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structure of the High-Salt Form of the Peridinin-Chlorophyll A-Protein from the Dinoflagellate Amphidinium Carterae: Modulation of the Spectral Properties of Pigments by the Protein Environment.
Biochemistry, 48, 2009
|
|
5V2C
 
 | | RE-REFINEMENT OF CRYSTAL STRUCTURE OF PHOTOSYSTEM II COMPLEX | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, J, Wiwczar, J.M, Brudvig, G.W. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlorophyll a with a farnesyl tail in thermophilic cyanobacteria.
Photosyn. Res., 134, 2017
|
|
5V6B
 
 | | Crystal structure of GIPC1 | | Descriptor: | PDZ domain-containing protein GIPC1 | | Authors: | Shang, G, Zhang, X. | | Deposit date: | 2017-03-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex.
Elife, 6, 2017
|
|
5UZZ
 
 | |
1J4P
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T155) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
5TFD
 
 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TNO
 
 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, SODIUM ION, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, H, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
