7NIJ
 
 | |
7NLV
 
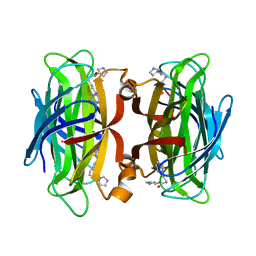 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE II | | Descriptor: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, Streptavidin | | Authors: | Nodling, A.R, Santi, N, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
7VT1
 
 | |
6XS9
 
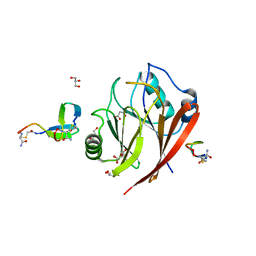 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L1 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 48V-TYR-ILE-LYS-THR-PRO-LEU-GLY-THR-PHE-PRO-ASN-ARG-HIS-GLY, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
7PO2
 
 | | Initiation complex of human mitochondrial ribosome small subunit with IF2, fMet-tRNAMet and mRNA | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7VRS
 
 | | The complex of Acyltransferase and Acyl Carrier Protein Domains from module 9 of Salinomycin Polyketide Synthase | | Descriptor: | 1,1'-butane-1,4-diylbis(1H-pyrrole-2,5-dione), 4'-PHOSPHOPANTETHEINE, Type I modular polyketide synthase | | Authors: | Feng, Y, Zheng, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural visualization of transient interactions between the cis-acting acyltransferase and acyl carrier protein of the salinomycin modular polyketide synthase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PNX
 
 | | Assembly intermediate of human mitochondrial ribosome small subunit without mS37 in complex with RBFA and METTL15 conformation a | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7PV1
 
 | |
1P7H
 
 | | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kB element | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*A)-3', 5'-D(*TP*TP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Giffin, M.J, Stroud, J.C, Bates, D.L, von Koenig, K.D, Hardin, J, Chen, L. | | Deposit date: | 2003-05-01 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element
Nat.Struct.Biol., 10, 2003
|
|
4XJ7
 
 | |
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
5OUN
 
 | | NMR solution structure of the external DII domain of Rvb2 from Saccharomyces cerevisiae | | Descriptor: | RuvB-like protein 2 | | Authors: | Rouillon, C, Bragantini, B, Charpentier, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-08-24 | | Release date: | 2018-03-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR assignment and solution structure of the external DII domain of the yeast Rvb2 protein.
Biomol NMR Assign, 12, 2018
|
|
4QAQ
 
 | | 1.58 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
8D5N
 
 | | Crystal structure of Ld-HF10 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
8J51
 
 | |
8D5P
 
 | | Mouse TCR TG6 | | Descriptor: | TCR-alpha, TCR-beta | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
8J50
 
 | |
6XN4
 
 | | Structure of the Lactococcus lactis Csm CTR_3:2 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
7TF2
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) Q484I spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
8J52
 
 | | Crystal structure of Flavihumibacter petaseus GH31 alpha-galactosidase mutant D304A in complex with alpha-1,4-galactobiose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GH31 alpha-galactosidase, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analysis of bacterial GH31 alpha-galactosidases specific for alpha-(1→4)-galactobiose.
Febs J., 290, 2023
|
|
6XN5
 
 | | Structure of the Lactococcus lactis Csm Apo- CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm3, CRISPR-associated protein Csm4, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN3
 
 | | Structure of the Lactococcus lactis Csm CTR_4:3 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
8J53
 
 | |
6XN7
 
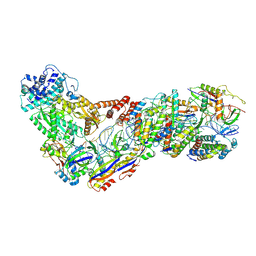 | | Structure of the Lactococcus lactis Csm NTR CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
7TF0
 
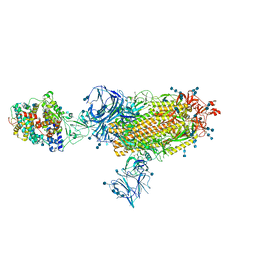 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
