5O7X
 
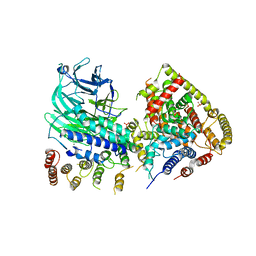 | | CRYSTAL STRUCTURE OF S. CEREVISIAE CORE FACTOR AT 3.2A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA polymerase I-specific transcription initiation factor RRN11, RNA polymerase I-specific transcription initiation factor RRN6, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-06-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
5ZOD
 
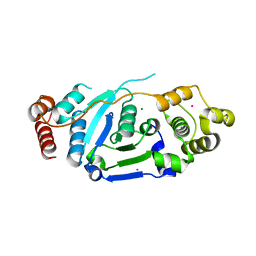 | | Crystal Structure of hFen1 in apo form | | Descriptor: | Flap endonuclease 1, MAGNESIUM ION, POTASSIUM ION | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
4FF4
 
 | |
2JAZ
 
 | |
1ORC
 
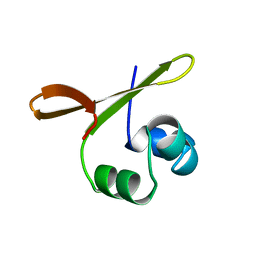 | |
8Q5N
 
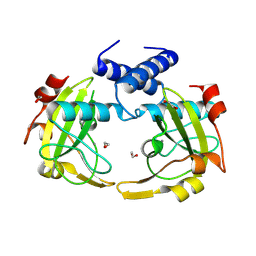 | | Apo form of restriction endonuclease NhoI. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Restriction endonuclease (NhoI) | | Authors: | Rafalski, D, Krakowska, A, Bochtler, M. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of the BisI family of modification dependent restriction endonucleases.
Nucleic Acids Res., 52, 2024
|
|
1OQJ
 
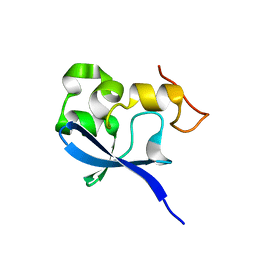 | | Crystal structure of the SAND domain from glucocorticoid modulatory element binding protein-1 (GMEB1) | | Descriptor: | Glucocorticoid Modulatory Element Binding protein-1, ZINC ION | | Authors: | Surdo, P.L, Bottomley, M.J, Sattler, M, Scheffzek, K. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions
MOL.ENDOCRINOL., 17, 2003
|
|
5JEM
 
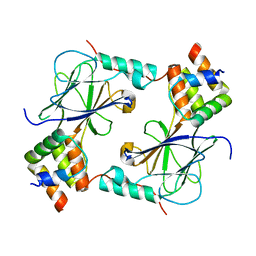 | | Complex of IRF-3 with CBP | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4IQU
 
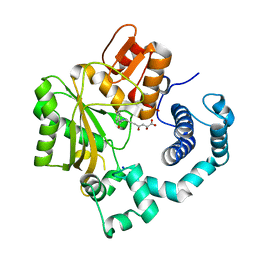 | | Tdt core in complex with inhibitor (2Z,5E)-6-[4-(4-fluorobenzoyl)-1H-pyrrol-2-yl]-2-hydroxy-4-oxohexa-2,5-dienoic acid | | Descriptor: | (2Z,5E)-6-[4-(4-fluorobenzoyl)-1H-pyrrol-2-yl]-2-hydroxy-4-oxohexa-2,5-dienoic acid, DNA nucleotidylexotransferase, SODIUM ION | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2013-01-13 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New nucleotide-competitive non-nucleoside inhibitors of terminal deoxynucleotidyl transferase: discovery, characterization, and crystal structure in complex with the target.
J.Med.Chem., 56, 2013
|
|
3HRM
 
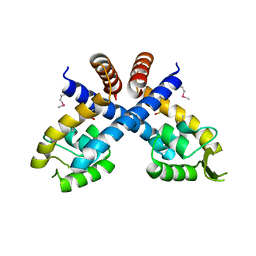 | | Crystal structure of Staphylococcus aureus protein SarZ in sulfenic acid form | | Descriptor: | HTH-type transcriptional regulator sarZ | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
1U94
 
 | |
5JEL
 
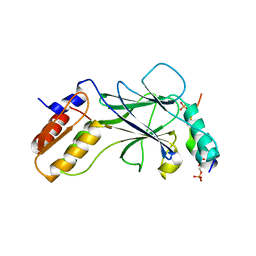 | | Phosphorylated TRIF in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEO
 
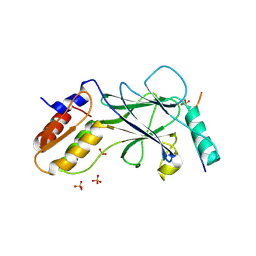 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5Y81
 
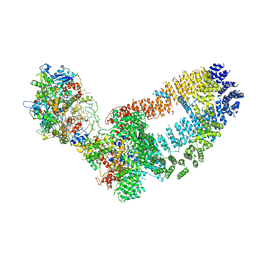 | | NuA4 TEEAA sub-complex | | Descriptor: | Actin, Actin-related protein 4, Chromatin modification-related protein EAF1, ... | | Authors: | Wang, X, Cai, G. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex
Nat Commun, 9, 2018
|
|
2N2C
 
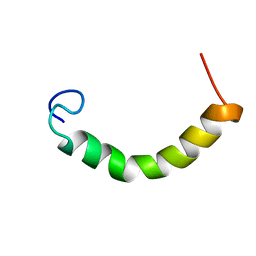 | | NMR Structure of TDP-43 prion-like hydrophobic helix in DPC | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Lim, L, Song, J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ALS-causing mutations significantly perturb the self-assembly and interaction with nucleic acid of the intrinsically-disordered prion-like domain of TDP-43
To be Published
|
|
6BDA
 
 | | Wild-type I-OnuI bound to A3G substrate (post-cleavage complex) | | Descriptor: | Cleaved Cognate DNA strand, +11 sense, Cleaved cognate DNA strand, ... | | Authors: | Brown, C, Zhang, K, Laforet, M, McMurrough, T, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Wild-type I-OnuI bound to A3G substrate (post-cleavage complex)
To Be Published
|
|
5Y7Y
 
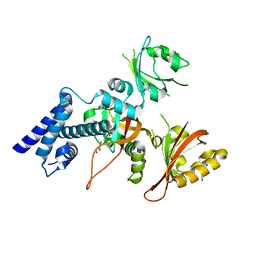 | | Crystal structure of AhRR/ARNT complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Aryl hydrocarbon receptor repressor, GLYCEROL | | Authors: | Sakurai, S, Shimizu, T, Ohto, U. | | Deposit date: | 2017-08-18 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AhRR-ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription.
J. Biol. Chem., 292, 2017
|
|
1U99
 
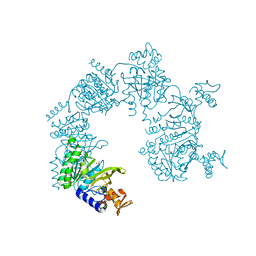 | |
4A63
 
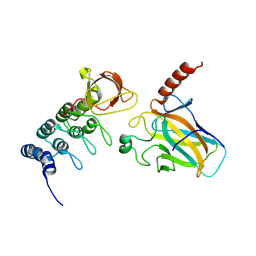 | | Crystal structure of the p73-ASPP2 complex at 2.6A resolution | | Descriptor: | ACETATE ION, APOPTOSIS STIMULATING OF P53 PROTEIN 2, TUMOUR PROTEIN 73, ... | | Authors: | Canning, P, Sharpe, T, Krojer, T, Savitsky, P, Cooper, C.D.O, Salah, E, Keates, T, Muniz, J, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C, Bountra, C, Edwards, A, Bullock, A.N. | | Deposit date: | 2011-10-31 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
5JER
 
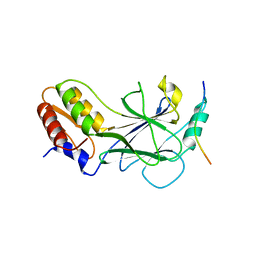 | | Structure of Rotavirus NSP1 bound to IRF-3 | | Descriptor: | Interferon regulatory factor 3, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1NUO
 
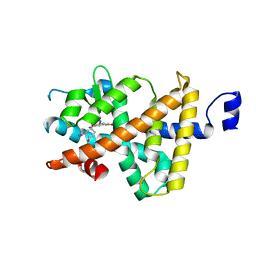 | | Two RTH Mutants with Impaired Hormone Binding | | Descriptor: | Thyroid hormone receptor beta-1, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Huber, B.R, Sandler, B, West, B.L, Cunha-Lima, S.T, Nguyen, H.T, Apriletti, J.W, Baxter, J.D, Fletterick, R.J. | | Deposit date: | 2003-01-31 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Two resistance to thyroid hormone mutants with impaired hormone binding
Mol.Endocrinol., 17, 2003
|
|
6WO0
 
 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | Descriptor: | GLYCEROL, Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
8BQS
 
 | | Cryo-EM structure of the I-II-III2-IV2 respiratory supercomplex from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mitochondrial membrane bending by I-II-III2-IV2 supercomplex
To Be Published
|
|
6WNL
 
 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | Descriptor: | Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
5JEK
 
 | | Phosphorylated MAVS in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, MAVS peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
