6CPB
 
 | |
6TT5
 
 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6QLE
 
 | | Structure of inner kinetochore CCAN complex | | Descriptor: | Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3,Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3, Central kinetochore subunit MCM16,Central kinetochore subunit MCM16,Inner kinetochore subunit MCM16,Mcm16p, Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
3JTZ
 
 | | Structure of the arm-type binding domain of HPI integrase | | Descriptor: | Integrase, SODIUM ION | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
6QLF
 
 | | Structure of inner kinetochore CCAN complex with mask1 | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6WO0
 
 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | Descriptor: | GLYCEROL, Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
3JU0
 
 | | Structure of the arm-type binding domain of HAI7 integrase | | Descriptor: | Phage integrase | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
2ZDS
 
 | | Crystal Structure of SCO6571 from Streptomyces coelicolor A3(2) | | Descriptor: | Putative DNA-binding protein | | Authors: | Begum, P, Gao, Y.G, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SCO6571 from Streptomyces coelicolor A3(2).
Protein Pept.Lett., 15, 2008
|
|
3T8R
 
 | | Crystal structure of Staphylococcus aureus CymR | | Descriptor: | Staphylococcus aureus CymR | | Authors: | He, C, Ji, Q. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
6WNL
 
 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | Descriptor: | Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
3P56
 
 | |
5DAJ
 
 | | Crystal structure of NalD, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | Descriptor: | NalD | | Authors: | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa
Mol.Microbiol., 100, 2016
|
|
4MH8
 
 | |
5Z30
 
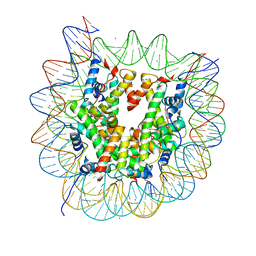 | | The crystal structure of the nucleosome containing a cancer-associated histone H2A.Z R80C mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.Z, ... | | Authors: | Horikoshi, N, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
2ORC
 
 | |
3T8T
 
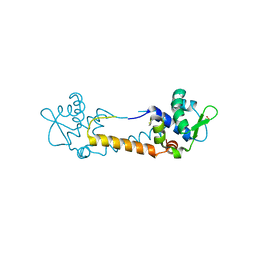 | | Crystal structure of Staphylococcus aureus CymR oxidized form | | Descriptor: | Staphylococcus aureus CymR (oxidized form) | | Authors: | He, C, Ji, Q. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
3ER8
 
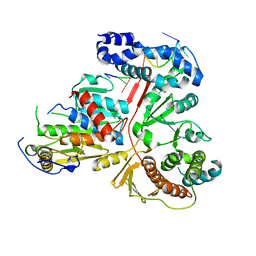 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with two fragments of RNA | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit, RNA/DNA chimera 5'-D(CP*CP*)R(UP*UP*)D(C)-3', ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
4EI7
 
 | | Crystal structure of Bacillus cereus TubZ, GDP-form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Plasmid replication protein RepX | | Authors: | Hayashi, I, Hoshino, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid.
J.Biol.Chem., 287, 2012
|
|
4EI9
 
 | | Crystal structure of Bacillus cereus TubZ, GTP-form | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Plasmid replication protein RepX | | Authors: | Hayashi, I, Hoshino, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid.
J.Biol.Chem., 287, 2012
|
|
4EGC
 
 | |
4EI8
 
 | |
4G06
 
 | | Crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae complexed with ssDNA. Northeast Structural Genomics Consortium (NESG) target SPR104 | | Descriptor: | DI(HYDROXYETHYL)ETHER, THYMIDINE-5'-MONOPHOSPHATE, Uncharacterized protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
8TFG
 
 | |
1A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-04-15 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
7AGI
 
 | | The structure of Artemis variant H35D | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
