8G2W
 
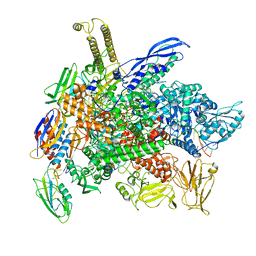 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G8Z
 
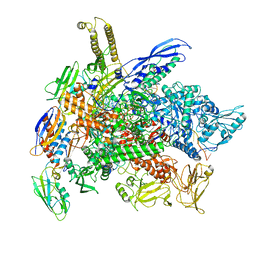 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-MER), ... | | Authors: | Porta, J.C, Ohi, M.D, Walter, N.G, Frank, A.T, Deb, I, Meze, K. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G1S
 
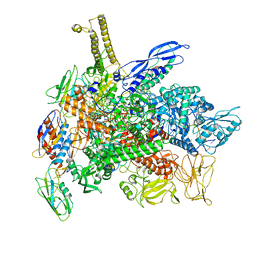 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G00
 
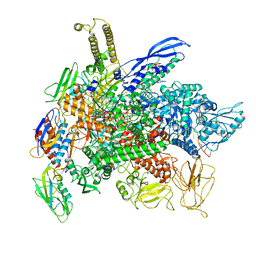 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-mer), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G7E
 
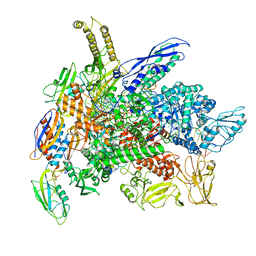 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2JO9
 
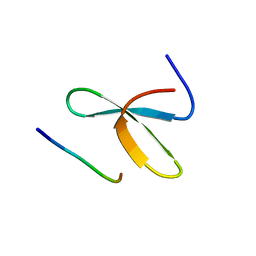 | | Mouse Itch 3rd WW domain complex with the Epstein-Barr virus latent membrane protein 2A derived peptide EEPPPPYED | | Descriptor: | Itchy E3 ubiquitin protein ligase, Latent membrane protein 2 | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
1R1K
 
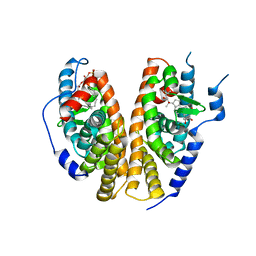 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.-M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-24 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
7WJ4
 
 | |
7WIZ
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, GLUTAMINE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liu, J.L, Guo, C.J. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1JW4
 
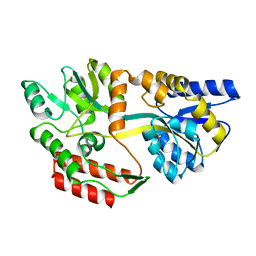 | | Structure of ligand-free maltodextrin-binding protein | | Descriptor: | maltodextrin-binding protein | | Authors: | Duan, X, Quiocho, F.A. | | Deposit date: | 2001-09-02 | | Release date: | 2002-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands.
Biochemistry, 41, 2002
|
|
4CPF
 
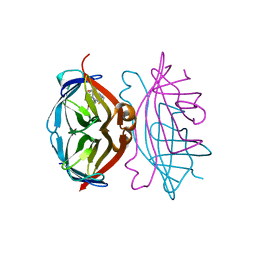 | | Wild-type streptavidin in complex with love-hate ligand 3 (LH3) | | Descriptor: | STREPTAVIDIN, methyl 4-(2-{5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H- thieno[3,4-d]imidazolidin-4-yl]pentanehydrazido}-3- [4-(methoxycarbonyl)phenyl]phenyl)benzoate | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
1D2R
 
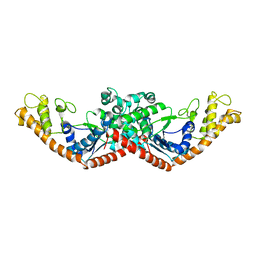 | |
4W9I
 
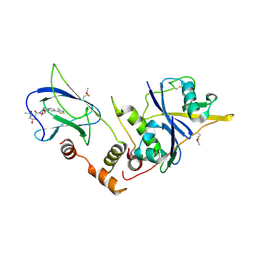 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((2S,4R)-1-acetyl-4-hydroxypyrrolidine-2-carbonyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 10) | | Descriptor: | (4R)-1-acetyl-4-hydroxy-L-prolyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4CPI
 
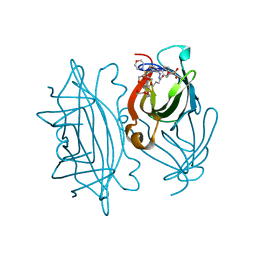 | | streptavidin A86D mutant with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4CPE
 
 | | Wild-type streptavidin in complex with love-hate ligand 1 (LH1) | | Descriptor: | (3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N-{2-[(2,6- diphenylphenyl)formamido]ethyl}pentanamide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4W9C
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
1K74
 
 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K7L
 
 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4W9D
 
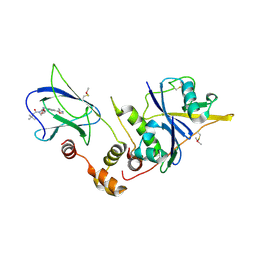 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methyloxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9H
 
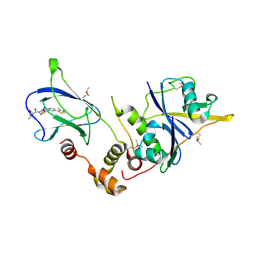 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-acetamido-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 7) | | Descriptor: | N-acetyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
8F3C
 
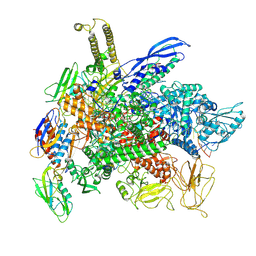 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4ZNV
 
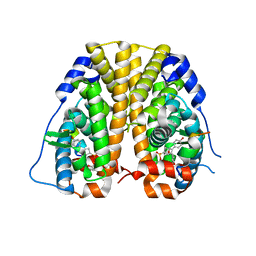 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Methoxy-substituted OBHS derivative | | Descriptor: | 2-methoxyphenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNS
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Fluoro-substituted OBHS derivative | | Descriptor: | 3-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7PT0
 
 | | SCO3201 with putative ligand | | Descriptor: | SPERMIDINE, TetR family transcriptional regulator | | Authors: | Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2021-09-25 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of free and ligand-bound forms of the TetR/AcrR-like regulator SCO3201 from Streptomyces coelicolor suggest a novel allosteric mechanism.
Febs J., 290, 2023
|
|
4ZNU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Methyl-substituted OBHS derivative | | Descriptor: | 2-methylphenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
