7KIB
 
 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 4 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Palte, R.L, Hayes, R.P. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7BV7
 
 | | INTS3 complexed with INTS6 | | Descriptor: | Integrator complex subunit 3, Integrator complex subunit 6 | | Authors: | Jia, Y, Bharath, S.R, Song, H. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair.
Cell Discov, 7, 2021
|
|
6Y6X
 
 | | Tetracenomycin X bound to the human ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel.
Nat.Chem.Biol., 16, 2020
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6YVV
 
 | | Condensin complex from S.cerevisiae ATP-free apo bridged state | | Descriptor: | Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Haering, C.H, Aragon, L, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8QH5
 
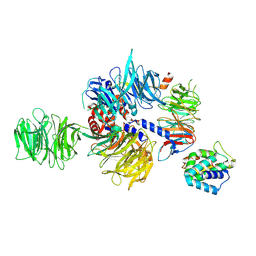 | | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1 | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2023-09-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
To Be Published
|
|
6N2G
 
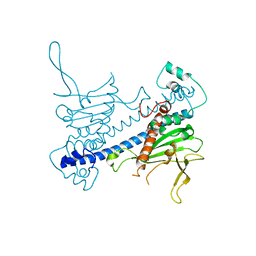 | | Crystal structure of Caenorhabditis elegans NAP1 | | Descriptor: | Nucleosome Assembly Protein | | Authors: | Bhattacharyya, S, DArcy, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Characterization of Caenorhabditis elegans Nucleosome Assembly Protein 1 Uncovers the Role of Acidic Tails in Histone Binding.
Biochemistry, 58, 2019
|
|
2E1N
 
 | |
1H56
 
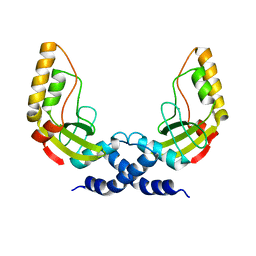 | | Structural and biochemical characterization of a new magnesium ion binding site near Tyr94 in the restriction endonuclease PvuII | | Descriptor: | MAGNESIUM ION, TYPE II RESTRICTION ENZYME PVUII | | Authors: | Spyrida, A, Matzen, C, Lanio, T, Jeltsch, A, Simoncsits, A, Athanasiadis, A, Scheuring-Vanamee, E, Kokkinidis, M, Pingoud, A. | | Deposit date: | 2001-05-20 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Characterization of a New Mg(2+) Binding Site Near Tyr94 in the Restriction Endonuclease PvuII.
J.Mol.Biol., 331, 2003
|
|
2GKD
 
 | |
1GRJ
 
 | |
2F3X
 
 | |
4X23
 
 | |
9J1S
 
 | |
4XLG
 
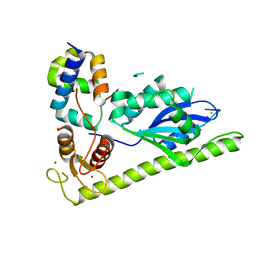 | | C. glabrata Slx1 in complex with Slx4CCD. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ... | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
8UN9
 
 | |
2HTJ
 
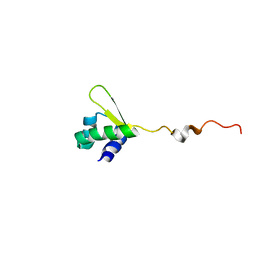 | | NMR structure of E.coli PapI | | Descriptor: | P fimbrial regulatory protein KS71A | | Authors: | Kawamura, T, Zhou, H, Le, L.U.K, Dahlquist, F.W. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Escherichia coli PapI, a Key Regulator of the Pap Pili Phase Variation.
J.Mol.Biol., 365, 2007
|
|
8TK0
 
 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4O5V
 
 | | Crystal structure of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8UV1
 
 | | Structure of TDP1 complexed with compound IB01 | | Descriptor: | 1,2-ETHANEDIOL, 8-(fluorosulfonyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke, T.R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of TDP1 complexed with compound IB01
To Be Published
|
|
8UZV
 
 | | Structure of TDP1 catalytic domain complexed with compound IB02 | | Descriptor: | 1,2-ETHANEDIOL, 8-{[2-(fluorosulfonyl)ethyl]amino}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Structure of TDP1 catalytic domain complexed with compound IB02
To Be Published
|
|
8UZZ
 
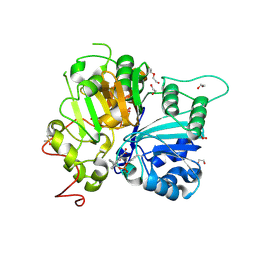 | | Structure of TDP1 catalytic domain complexed with compound IB03 | | Descriptor: | (8M)-8-{2-[(fluorosulfonyl)oxy]phenyl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structureal analysis of TDP1 in complex with inhbitors
To Be Published
|
|
8V0C
 
 | | Structure of TDP1 catalytic domain complexed with compound IB06 | | Descriptor: | (8M)-8-(2-{[2-(fluorosulfonyl)ethyl]amino}phenyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of TDP1 complexed with inhibitors
To Be Published
|
|
8V0B
 
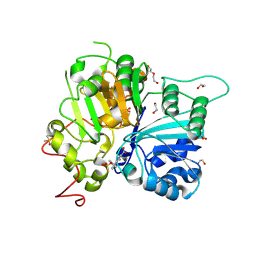 | | Structure of TDP1 catalytic domain complexed with compound IB05 | | Descriptor: | 1,2-ETHANEDIOL, 8-{4-[(fluorosulfonyl)oxy]phenyl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of TDP1 complexed with inhibitors
To Be Published
|
|
8TWH
 
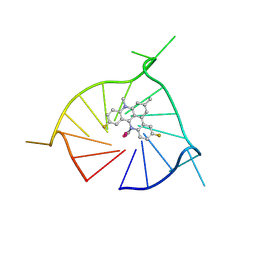 | | Crystal structure of (GGGTT)3GGG G-quadruplex in complex with small molecule ligand RHPS4 | | Descriptor: | (GGGTT)3GGG DNA, 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, POTASSIUM ION | | Authors: | Yatsunyk, L.A, Martin, K.N, Lam, G. | | Deposit date: | 2023-08-21 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of (GGGTT)3GGG G-quadruplex in complex with small molecule ligand RHPS4
To be published
|
|
