2B5D
 
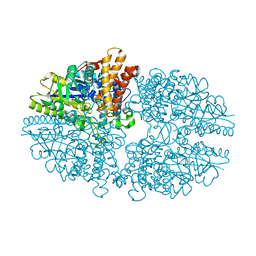 | | Crystal structure of the novel alpha-amylase AmyC from Thermotoga maritima | | Descriptor: | alpha-Amylase | | Authors: | Dickmanns, A, Ballschmiter, M, Liebl, W, Ficner, R. | | Deposit date: | 2005-09-28 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the novel alpha-amylase AmyC from Thermotoga maritima.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IJ2
 
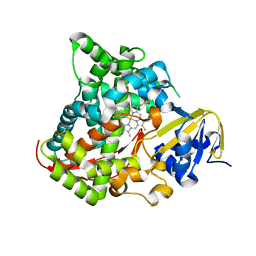 | | Atomic structure of the heme domain of flavocytochrome P450-BM3 | | Descriptor: | Cytochrome P450 BM3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Helen, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
6XKO
 
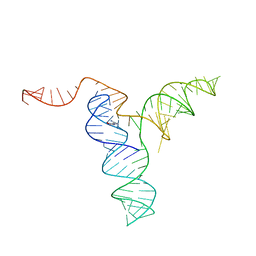 | |
2BA0
 
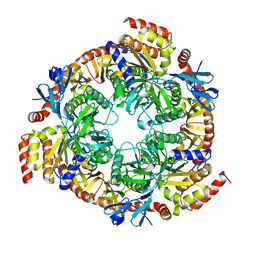 | | Archaeal exosome core | | Descriptor: | Archaeal exosome RNA binding protein RRP4, Archaeal exosome RNA binding protein RRP41, Archaeal exosome RNA binding protein RRP42 | | Authors: | Buttner, K, Wenig, K, Hopfner, K.P. | | Deposit date: | 2005-10-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural framework for the mechanism of archaeal exosomes in RNA processing.
Mol.Cell, 20, 2005
|
|
2IC6
 
 | |
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | Descriptor: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
6Y67
 
 | |
7N84
 
 | | Double nuclear outer ring from the isolated yeast NPC | | Descriptor: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N3L
 
 | | Co-complex CYP46A1 with 0420 (compound 6) | | Descriptor: | 1,2-ETHANEDIOL, Cholesterol 24-hydroxylase, GLYCEROL, ... | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of Novel 3-Piperidinyl Pyridine Derivatives as Highly Potent and Selective Cholesterol 24-Hydroxylase (CH24H) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7N3M
 
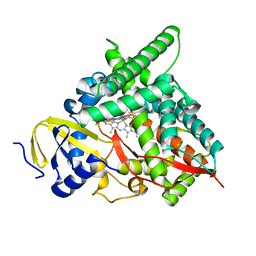 | | Co-complex CYP46A1 with 0431 (compound 17) | | Descriptor: | Cholesterol 24-hydroxylase, N,N-dimethyl-1-[4-(4-methyl-1H-pyrazol-1-yl)pyridin-3-yl]piperidine-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Discovery of Novel 3-Piperidinyl Pyridine Derivatives as Highly Potent and Selective Cholesterol 24-Hydroxylase (CH24H) Inhibitors.
J.Med.Chem., 65, 2022
|
|
6Y7F
 
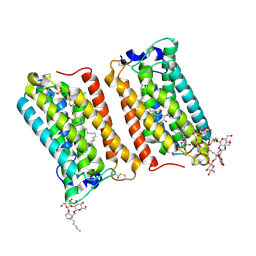 | | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7) | | Descriptor: | CHLORIDE ION, Elongation of very long chain fatty acids protein 7, Octyl Glucose Neopentyl Glycol, ... | | Authors: | Nie, L, Pike, A.C.W, Bushell, S.R, Chu, A, Cole, V, Speedman, D, Rodstrom, K.E.J, Kupinska, K, Shrestha, L, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7)
TO BE PUBLISHED
|
|
7NE3
 
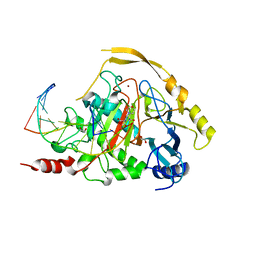 | | Human TET2 in complex with favourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
2ID9
 
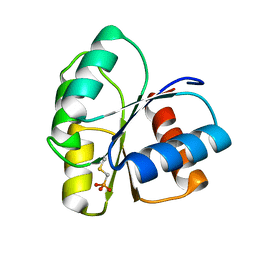 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
6YFU
 
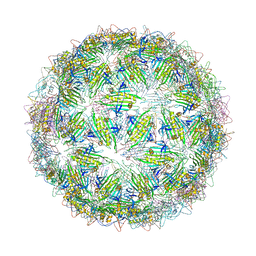 | | Virus-like particle of Wenzhou levi-like virus 4 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.018 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
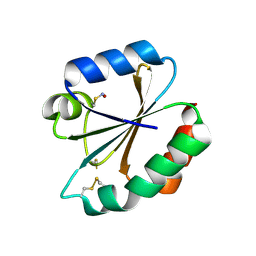
2IFQ
 
 | |
2J45
 
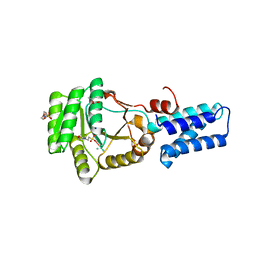 | | Water structure of T. Aquaticus Ffh NG Domain At 1.1A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Freymann, D.M, Ramirez, U.D. | | Deposit date: | 2006-08-24 | | Release date: | 2006-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Analysis of Protein Hydration in Ultra-High Resolution Structures of the Srp Gtpase Ffh
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2J9J
 
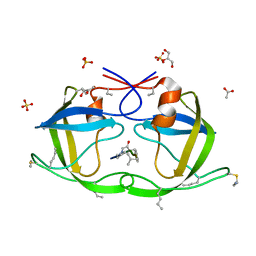 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
6YAI
 
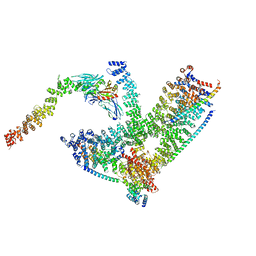 | | Clathrin with bound beta2 appendage of AP2 | | Descriptor: | AP-2 complex subunit beta, Clathrin heavy chain, Clathrin light chain | | Authors: | Kovtun, O, Kane Dickson, V, Kelly, B.T, Owen, D, Briggs, J.A.G. | | Deposit date: | 2020-03-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
2JDX
 
 | |
6YF7
 
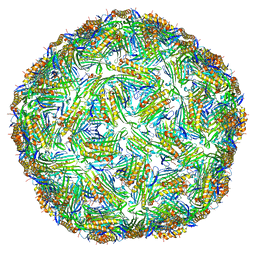 | | Virus-like particle of bacteriophage AC | | Descriptor: | Coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
7N5V
 
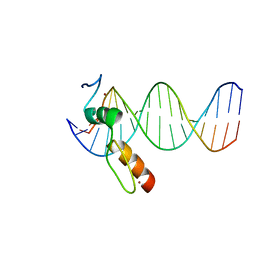 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
2JE4
 
 | | Atomic-resolution crystal structure of chemically-synthesized HIV-1 protease in complex with JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
J.Am.Chem.Soc., 129, 2007
|
|
7N4E
 
 | | Escherichia coli sigma 70-dependent paused transcription elongation complex | | Descriptor: | 5'-R(*UP*UP*CP*GP*GP*AP*GP*AP*GP*GP*UP*A)-3', DNA (61-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Su, M, Ebright, R.H. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic basis of sigma-dependent transcriptional pausing.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2JJ1
 
 | | The Structure of F1-ATPase inhibited by piceatannol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
