5MMB
 
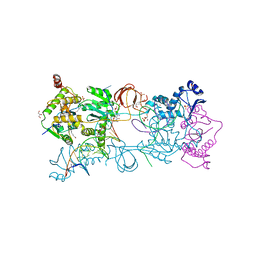 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ434 (compound 6p) | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), GLYCEROL, ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors.
J. Med. Chem., 60, 2017
|
|
8XYE
 
 | | Crystal structure of SARS-CoV-2 BA.4 RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Lan, J, Wang, C.H. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Receptor binding mechanism and immune evasion capacity of SARS-CoV-2 BQ.1.1 lineage.
Virology, 600, 2024
|
|
1N5S
 
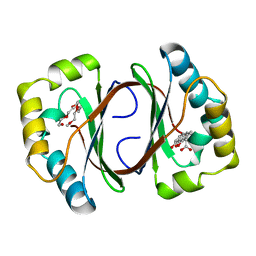 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9-OXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5T
 
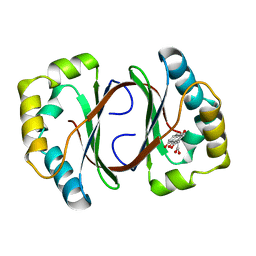 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Oxidized Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9,10-DIOXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, G Kendrew, S, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
8GC6
 
 | |
8GDV
 
 | |
5GX8
 
 | | Crystal structure of solute-binding protein related to glycosaminoglycans | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
6QC4
 
 | | Ovine respiratory supercomplex I+III2 open class 3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Acyl carrier protein, Cytochrome b, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6H52
 
 | | Crystal structure of human KDM5B in complex with compound 34g | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(1-cyclopentylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
5TZQ
 
 | | Crystal Structure of FPV039:Bmf BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein FPV039, Bcl-2-modifying factor | | Authors: | Anasir, M.I, Kvansakul, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J. Biol. Chem., 292, 2017
|
|
5U26
 
 | |
5E78
 
 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
6H51
 
 | | Crystal structure of human KDM5B in complex with compound 34f | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[1-(cyclobutylmethyl)piperidin-4-yl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6HFW
 
 | | Mycobacterium tuberculosis DprE1 in complex with CMP1 | | Descriptor: | 8-(oxidanylamino)-2-piperidin-1-yl-6-(trifluoromethyl)-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chung, C. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
4NBM
 
 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
5U9P
 
 | |
9JP2
 
 | | COMPLEX STRUCTURE OF ENDO-1,3-FUCANASE WITH FUCOTETRAOSE | | Descriptor: | Glycoside-hydrolase family GH114 TIM-barrel domain-containing protein, alpha-L-fucopyranose-(1-3)-2,4-di-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose | | Authors: | Chen, G.N, Chang, Y.G. | | Deposit date: | 2024-09-25 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Complex structure of endo-1.3-fucanase with fucotetraose at 2.02 Angstroms resulution.
To Be Published
|
|
9K1G
 
 | |
6VSL
 
 | | Crystal structure of a human fucosylated IgG1 Fc expressed in tobacco plants (Nicotiana benthamiana) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tolbert, W.D, Pazgier, M. | | Deposit date: | 2020-02-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced Ability of Plant-Derived PGT121 Glycovariants To Eliminate HIV-1-Infected Cells.
J.Virol., 95, 2021
|
|
5NAW
 
 | | Complement factor D in complex with the inhibitor (1R,3S,5R)-2-Aza-bicyclo[3.1.0]hexane-2,3-dicarboxylic acid 2-[(1-carbamoyl-1H-indol-3-yl)-amide] 3-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (1~{R},3~{S},5~{R})-~{N}2-(1-aminocarbonylindol-3-yl)-~{N}3-[3-(trifluoromethyloxy)phenyl]-2-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NKA
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2g | | Descriptor: | 1,2-ETHANEDIOL, 4-[[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylamino]cyclohexane-1-carboxylic acid, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
9FDS
 
 | | Crystal structure of human Sirt2 in complex with SirReal2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, CHLORIDE ION, ... | | Authors: | Friedrich, F, Einsle, O, Jung, M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Efficient Crystallization of Apo Sirt2 for Small-Molecule Soaking and Structural Analysis of Ligand Interactions.
J.Med.Chem., 68, 2025
|
|
8ZDX
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for human DPP4 receptor recognition by a pangolin MERS-like coronavirus.
Plos Pathog., 20, 2024
|
|
9FCP
 
 | | OPR3 loop swap variant L6(AchrOYE4) in complex with NADH4 | | Descriptor: | 1,4,5,6-Tetrahydronicotinamide adenine dinucleotide, 12-oxophytodienoate reductase 3,Old yellow enzyme OYE4, FLAVIN MONONUCLEOTIDE | | Authors: | Bijelic, A, Kerschbaumer, B, Macheroux, P. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the complexity of coenzyme specificity in ene-reductases
To Be Published
|
|
1LMI
 
 | | 1.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED PROTEIN FROM MYCOBACTERIUM TUBERCULOSIS-MPT63 | | Descriptor: | Immunogenic protein MPT63/MPB63 | | Authors: | Goulding, C.W, Parseghian, A, Sawaya, M.R, Cascio, D, Apostol, M, Gennaro, M.L, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-01 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at
1.5-A resolution
Protein Sci., 11, 2002
|
|
