2JQ2
 
 | |
3FAV
 
 | | Structure of the CFP10-ESAT6 complex from Mycobacterium tuberculosis | | Descriptor: | 6 kDa early secretory antigenic target, ESAT-6-like protein esxB, IMIDAZOLE, ... | | Authors: | Poulsen, C, Holton, S.J, Wilmanns, M, Song, Y.H. | | Deposit date: | 2008-11-18 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
4I5V
 
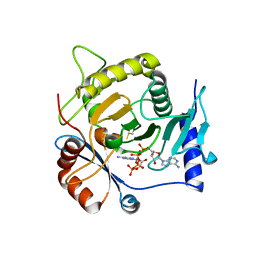 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with Ap4A | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, BIS(ADENOSINE)-5'-TETRAPHOSPHATE | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
4I5T
 
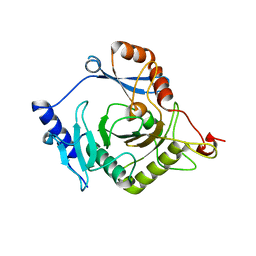 | | Crystal structure of yeast Ap4A phosphorylase Apa2 | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2 | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
4I5W
 
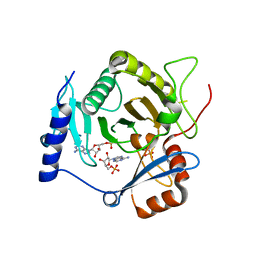 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with AMP | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, ADENOSINE MONOPHOSPHATE, PHOSPHATE ION | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
8HVQ
 
 | | Crystal structure of haloacid dehalogenase-like hydrolase family enzyme from Staphylococcus lugdunensis | | Descriptor: | 1,2-ETHANEDIOL, Cof-type HAD-IIB family hydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaur, H, Mahto, J.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Characterization of haloacid dehalogenase superfamily acid phosphatase from Staphylococcus lugdunensis.
Arch.Biochem.Biophys., 753, 2024
|
|
7OB3
 
 | | hSTING in complex with 3',3'-c-di-araAMP | | Descriptor: | 3',3'-c-di-araAMP, Stimulator of interferon genes protein | | Authors: | Smola, M, Boura, E. | | Deposit date: | 2021-04-20 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Synthesis of 3'-5', 3'-5' Cyclic Dinucleotides, Their Binding Properties to the Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations
Biochemistry, 2021
|
|
8D1V
 
 | | Cryo-EM structure of guide RNA and target RNA bound Cas7-11 | | Descriptor: | CRISPR RNA (34-MER), CRISPR-associated RAMP family protein, SS target RNA (5'-R(P*AP*GP*CP*UP*UP*GP*GP*UP*UP*CP*AP*AP*AP*GP*AP*AP*CP*G)-3'), ... | | Authors: | Rai, J, Goswami, H, Li, H. | | Deposit date: | 2022-05-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Molecular mechanism of active Cas7-11 in processing CRISPR RNA and interfering target RNA.
Elife, 11, 2022
|
|
3FYY
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with Mg | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
3M45
 
 | | Crystal structure of Ig1 domain of mouse SynCAM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 2 | | Authors: | Yue, L, Modis, Y. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion.
J.Biol.Chem., 285, 2010
|
|
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3VLC
 
 | |
3HPF
 
 | | Crystal structure of the mutant Y90F of divergent galactarate dehydratase from Oceanobacillus iheyensis complexed with Mg and galactarate | | Descriptor: | D-galactaric acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-06-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
2VNO
 
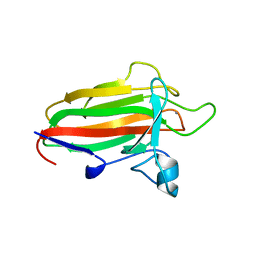 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens in complex with blood group B-trisaccharide ligand. | | Descriptor: | CALCIUM ION, CPE0329, alpha-L-fucopyranose-(1-2)-[beta-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
3GVM
 
 | |
8IK0
 
 | |
3GWK
 
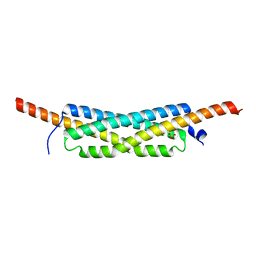 | | Structure of the homodimeric WXG-100 family protein from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein SAG1039, SULFATE ION | | Authors: | Poulsen, C, Gries, F, Wilmanns, M, Song, Y.H. | | Deposit date: | 2009-04-01 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
2OQY
 
 | | The crystal structure of muconate cycloisomerase from Oceanobacillus iheyensis | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Toro, R, Fedorov, E.V, Bonanno, J, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
2MZ8
 
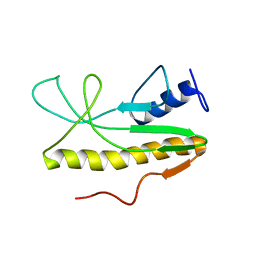 | | Solution NMR structure of Salmonella Typhimurium transcriptional regulator protein Crl | | Descriptor: | Sigma factor-binding protein Crl | | Authors: | Cavaliere, P, Levi-Acobas, F, Monteil, V, Bellalou, J, Mayer, C, Norel, F, Sizun, C. | | Deposit date: | 2015-02-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding interface between the Salmonella sigma (S)/RpoS subunit of RNA polymerase and Crl: hints from bacterial species lacking crl.
Sci Rep, 5, 2015
|
|
1NLM
 
 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | Descriptor: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2D1G
 
 | | Structure of Francisella tularensis Acid Phosphatase A (AcpA) bound to orthovanadate | | Descriptor: | 2-ETHOXYETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DECAVANADATE, ... | | Authors: | Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2005-08-20 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Francisella tularensis AcpA: prototype of a unique superfamily of acid phosphatases and phospholipases C
J.Biol.Chem., 281, 2006
|
|
