3SS9
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
1XC7
 
 | | Binding of beta-D-glucopyranosyl bismethoxyphosphoramidate to glycogen phosphorylase b: Kinetic and crystallographic studies | | Descriptor: | Glycogen phosphorylase, muscle form, N-(dimethoxyphosphoryl)-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Kardakaris, R, Bischler, N, Leonidas, D.D, Kannan, T, Loganathan, D, Oikonomakos, N.G. | | Deposit date: | 2004-09-01 | | Release date: | 2005-02-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Binding of beta-d-glucopyranosyl bismethoxyphosphoramidate to glycogen phosphorylase b: kinetic and crystallographic studies
Bioorg.Med.Chem., 13, 2005
|
|
1K06
 
 | | Crystallographic Binding Study of 100 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1E1F
 
 | | Crystal structure of a Monocot (Maize ZMGlu1) beta-glucosidase in complex with p-Nitrophenyl-beta-D-thioglucoside | | Descriptor: | 4-nitrophenyl 1-thio-beta-D-glucopyranoside, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-05-03 | | Release date: | 2001-02-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Monocotyledon (Maize Zmglu1) Beta-Glucosidase and a Model of its Complex with P-Nitrophenyl Beta-D-Thioglucoside
Biochem.J., 354, 2001
|
|
6I01
 
 | | Structure of human D-glucuronyl C5 epimerase in complex with substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I02
 
 | | Structure of human D-glucuronyl C5 epimerase in complex with product | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2Q3I
 
 | | Crystal structure of the D10-P3/IQN17 complex: a D-peptide inhibitor of HIV-1 entry bound to the GP41 coiled-coil pocket | | Descriptor: | CHLORIDE ION, D-peptide, Fusion protein between the Coiled-Coil pocket of HIV GP41 and gcn4-PIQI | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
1XL1
 
 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | (1R)-1,5-anhydro-1-(5-methyl-1,3-benzothiazol-2-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
1XL0
 
 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | (1R)-1,5-anhydro-1-(5-methyl-1,3,4-oxadiazol-2-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
1GDH
 
 | |
1FJA
 
 | |
1XKX
 
 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | 2-(BETA-D-GLUCOPYRANOSYL)-5-METHYL-1-BENZIMIDAZOLE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
1KTI
 
 | | BINDING OF 100 MM N-ACETYL-N'-BETA-D-GLUCOPYRANOSYL UREA TO GLYCOGEN PHOSPHORYLASE B: KINETIC AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-(acetylcarbamoyl)-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2002-01-16 | | Release date: | 2002-01-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of N-acetyl-N'-beta-D-glucopyranosyl urea and N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1GG8
 
 | | DESIGN OF INHIBITORS OF GLYCOGEN PHOSPHORYLASE: A STUDY OF ALPHA-AND BETA-C-GLUCOSIDES AND 1-THIO-BETA-D-GLUCOSE COMPOUNDS | | Descriptor: | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Watson, K.A, Mitchell, E.P, Johnson, L.N, Son, J.C, Bichard, C.J, Orchard, M.G, Fleet, G.W, Oikonomakos, N.G, Leonidas, D.D, Kontou, M, Papageorgiou, A.C. | | Deposit date: | 2000-07-30 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design of inhibitors of glycogen phosphorylase: a study of alpha- and beta-C-glucosides and 1-thio-beta-D-glucose compounds.
Biochemistry, 33, 1994
|
|
1DNK
 
 | | THE X-RAY STRUCTURE OF THE DNASE I-D(GGTATACC)2 COMPLEX AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*GP*GP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3'), ... | | Authors: | Weston, S.A, Lahm, A, Suck, D. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution.
J.Mol.Biol., 226, 1992
|
|
4AVV
 
 | | Structure of CPHPC bound to Serum Amyloid P Component | | Descriptor: | (2R)-1-[6-[(2R)-2-carboxypyrrolidin-1-yl]-6-oxidanylidene-hexanoyl]pyrrolidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kolstoe, S.E, Jenvey, M.C, Wood, S.P. | | Deposit date: | 2012-05-29 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of Serum Amyloid P Component with Hexanoyl Bis(D-Proline) (Cphpc)
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1CZQ
 
 | | CRYSTAL STRUCTURE OF THE D10-P1/IQN17 COMPLEX: A D-PEPTIDE INHIBITOR OF HIV-1 ENTRY BOUND TO THE GP41 COILED-COIL POCKET. | | Descriptor: | CHLORIDE ION, D-PEPTIDE INHIBITOR, FUSION PROTEIN BETWEEN THE HYDROPHOBIC POCKET OF HIV GP41 AND GCN4-PIQI | | Authors: | Eckert, D.M, Malashkevich, V.N, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 1999-09-06 | | Release date: | 1999-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket.
Cell(Cambridge,Mass.), 99, 1999
|
|
1K08
 
 | | Crystallographic Binding Study of 10 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1H8L
 
 | | Duck Carboxypeptidase D Domain II in complex with GEMSA | | Descriptor: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gomis-Rueth, F.X, Coll, M, Aviles, F.X, Vendrell, J, Fricker, L.D. | | Deposit date: | 2001-02-09 | | Release date: | 2002-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the inhibitor-complexed carboxypeptidase D domain II and the modeling of regulatory carboxypeptidases.
J. Biol. Chem., 276, 2001
|
|
2Q33
 
 | | Crystal structure of all-D monellin at 1.8 A resolution | | Descriptor: | D-MONELLIN CHAIN A, D-MONELLIN CHAIN B | | Authors: | Hung, L.-W, Kohmura, M, Ariyoshi, Y, Kim, S.-H. | | Deposit date: | 2007-05-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Enantiomeric Protein, D-Monellin at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1PX8
 
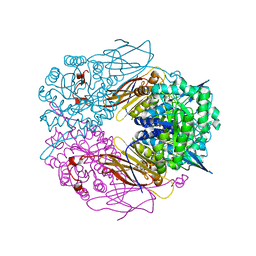 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
2HK0
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the absence of substrate | | Descriptor: | D-PSICOSE 3-EPIMERASE | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
2DKC
 
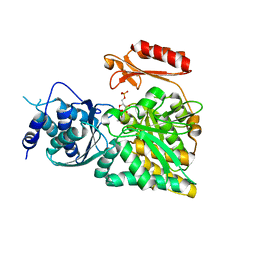 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the substrate complex | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2DKD
 
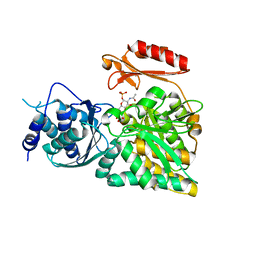 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
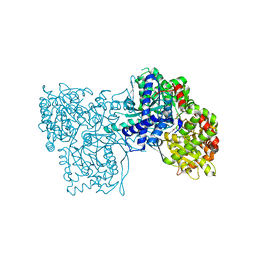
2F3U
 
 | |
