7QK7
 
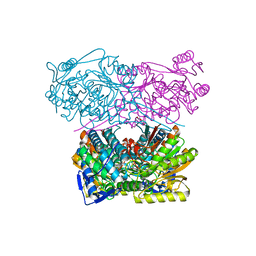 | | Crystal structure of the APO form of ALDH1A3 | | Descriptor: | Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
7QK9
 
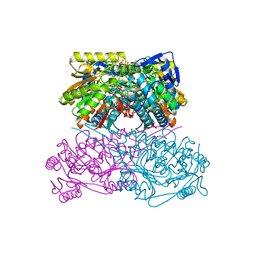 | | Crystal structure of the ALDH1A3-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
7QK8
 
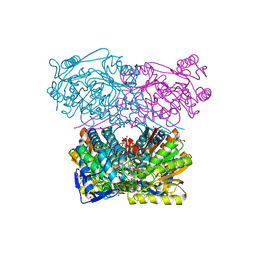 | | Crystal structure of the ALDH1A3-NAD+ complex | | Descriptor: | Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
8CGS
 
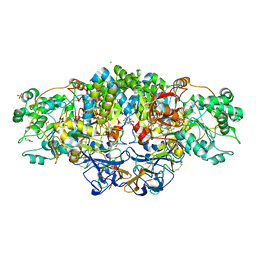 | | Crystal structure of arsenite oxidase from Alcaligenes faecalis (Af Aio) bound to antimony oxyanion | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ... | | Authors: | Engrola, F, Correia, M.A.S, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2023-02-06 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Arsenite oxidase in complex with antimonite and arsenite oxyanions: Insights into the catalytic mechanism.
J.Biol.Chem., 299, 2023
|
|
6VSG
 
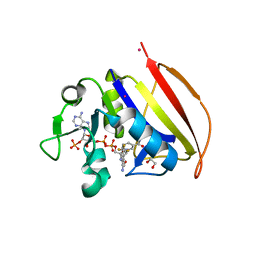 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 4-(trifluoromethyl)benzene-1,2-diamine(fragment 17) | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VSP
 
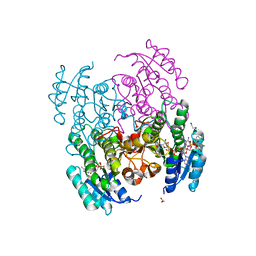 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase mutant Q247A | | Descriptor: | 1,2-ETHANEDIOL, 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
6YNF
 
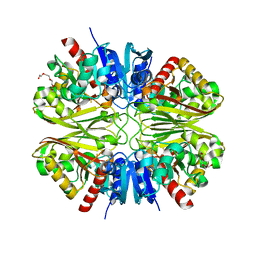 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 3 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6NP9
 
 | | PD-L1 IgV domain V76T with fragment | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6NOJ
 
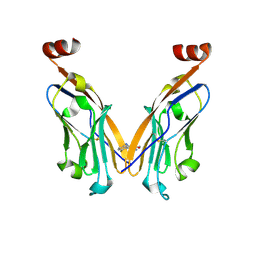 | | PD-L1 IgV domain V76T with fragment | | Descriptor: | Programmed cell death 1 ligand 1, methyl 3-amino-4-(2-fluorophenyl)-1H-pyrrole-2-carboxylate | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6VSF
 
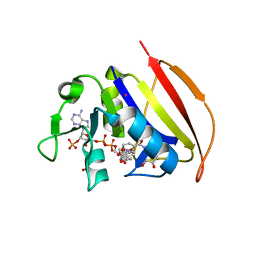 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 4-(3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-yl)-4-oxobutanoic acid(fragment 16) | | Descriptor: | 4-(3,4-dihydro-2H-1,5-benzodioxepin-7-yl)-4-oxobutanoic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6NPI
 
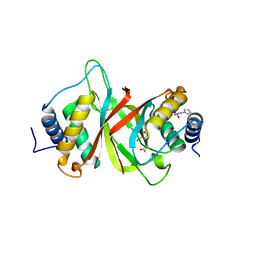 | | Crystal structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, bound to fragments | | Descriptor: | ({2-[(4-bromo-5-methyl-1,2-oxazol-3-yl)amino]-2-oxoethyl}sulfanyl)acetic acid, 2-pyrrol-1-ylbenzoic acid, Epstein-Barr nuclear antigen 1 | | Authors: | Messick, T.E. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth.
Sci Transl Med, 11, 2019
|
|
8YQE
 
 | | CDK2 and small molecular inhibitor L56 | | Descriptor: | 4-[[2,6-bis(fluoranyl)-3-(oxan-4-yl)phenyl]carbonylamino]-~{N}-(1-ethylsulfonylpiperidin-4-yl)-1~{H}-pyrazole-5-carboxamide, Cyclin-dependent kinase 2 | | Authors: | Ge, S. | | Deposit date: | 2024-03-19 | | Release date: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of CDK2 and L56 at 1.9 Angstorms resolution
To Be Published
|
|
6VW7
 
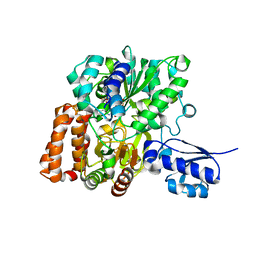 | | Formate Dehydrogenase FdsABG subcomplex FdsBG from C. necator - NADH bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Young, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and kinetic analyses of the FdsBG subcomplex of the cytosolic formate dehydrogenase FdsABG fromCupriavidus necator.
J.Biol.Chem., 295, 2020
|
|
8DFD
 
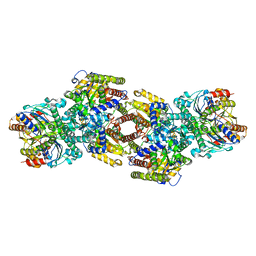 | |
7TQS
 
 | |
7TQT
 
 | |
4X3V
 
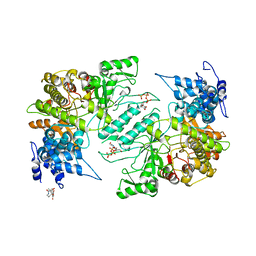 | | Crystal structure of human ribonucleotide reductase 1 bound to inhibitor | | Descriptor: | N~6~-{N-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetyl]-2-methyl-D-alanyl}-D-lysine, Ribonucleoside-diphosphate reductase large subunit, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Dealwis, C.G, Ahmad, M.F, Alam, I. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Identification of Non-nucleoside Human Ribonucleotide Reductase Modulators.
J.Med.Chem., 58, 2015
|
|
8T7N
 
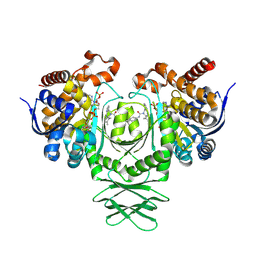 | | Crystal structure of the R132H mutant of IDH1 bound to compound 1 | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, N-(4-tert-butylphenyl)-7,8-dimethyl-5,11-dihydro-6H-pyrido[2,3-b][1,5]benzodiazepine-6-carboxamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lu, J, Abeywickrema, P, Heo, M.R, Parthasarathy, G, McCoy, M, Soisson, S.M. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biostructural, biochemical and biophysical studies of mutant IDH1.
Nat Commun, 15, 2024
|
|
6OME
 
 | |
4X9C
 
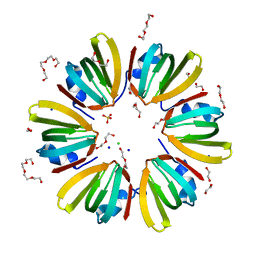 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
8U98
 
 | |
6O4I
 
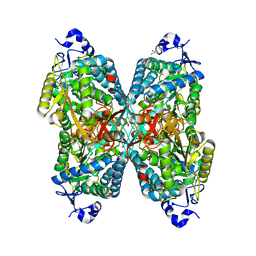 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
7RM6
 
 | |
8Z0E
 
 | |
6VS5
 
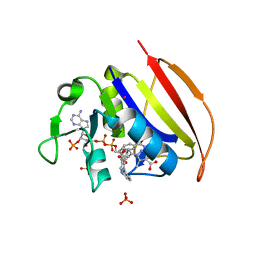 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 5-methyl-1-phenyl-1H-pyrazole-4-carboxylic acid (fragment 1) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Tyrakis, P, Blundell, T, Dias, M.V.B. | | Deposit date: | 2020-02-10 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
