5OF4
 
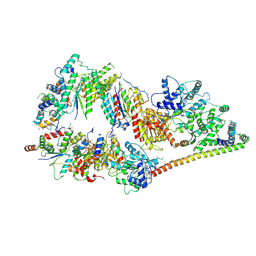 | | The cryo-EM structure of human TFIIH | | Descriptor: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, General transcription factor IIH subunit 4,p52,General transcription factor IIH subunit 4, ... | | Authors: | Greber, B.J, Nguyen, T.H.D, Fang, J, Afonine, P.V, Adams, P.D, Nogales, E. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The cryo-electron microscopy structure of human transcription factor IIH.
Nature, 549, 2017
|
|
2IOO
 
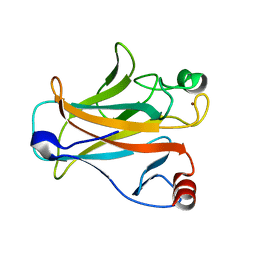 | | Crystal structure of the mouse p53 core domain | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4FXH
 
 | |
2IOI
 
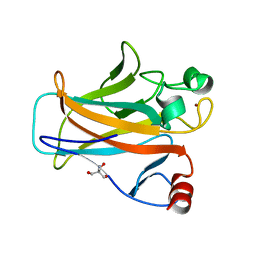 | | Crystal structure of the mouse p53 core domain at 1.55 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOM
 
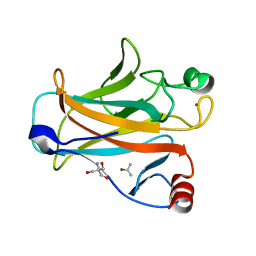 | | Mouse p53 core domain soaked with 2-propanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ISOPROPYL ALCOHOL, ... | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
5FHG
 
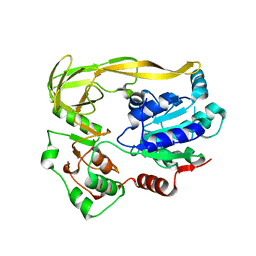 | |
4FXI
 
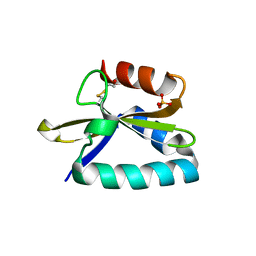 | |
7B48
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273H mutant bound to MQ: R273H-MQ (II) | | Descriptor: | (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Degtjarik, O, Rozenberg, H, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
8BAH
 
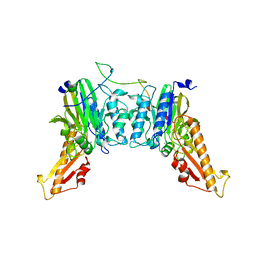 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
4K3Q
 
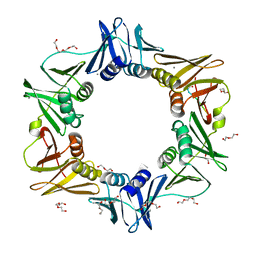 | | E. coli sliding clamp in complex with AcQLDAF | | Descriptor: | (ACE)QLDAF, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
7N40
 
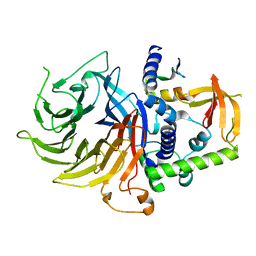 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
4H0E
 
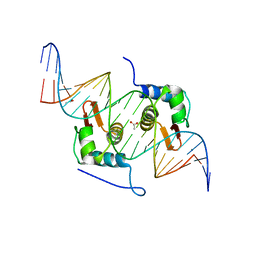 | | Crystal Structure of mutant ORR3 in complex with NTD of AraR | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*CP*CP*GP*TP*AP*CP*AP*TP*TP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*AP*AP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*AP*AP*TP*T)-3', ACETATE ION, ... | | Authors: | Nair, D.T, Jain, D. | | Deposit date: | 2012-09-08 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
1FC3
 
 | |
2E2J
 
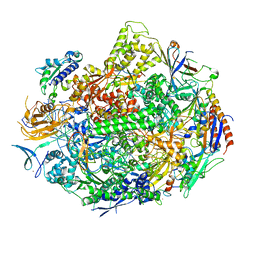 | | RNA polymerase II elongation complex in 5 mM Mg+2 with GMPCPP | | Descriptor: | 27-MER DNA template strand, 5'-D(P*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*A)-3', 5'-R(P*AP*UP*CP*GP*AP*GP*AP*GP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
5FGO
 
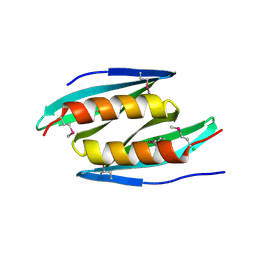 | |
2E2H
 
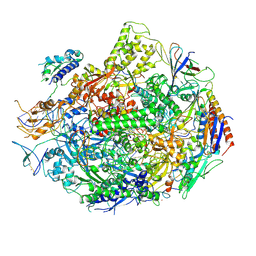 | | RNA polymerase II elongation complex at 5 mM Mg2+ with GTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2E2I
 
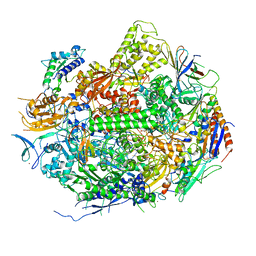 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
4K3R
 
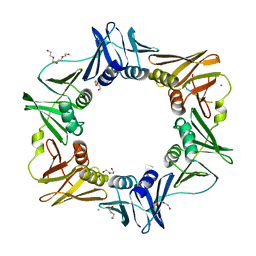 | | E. coli sliding clamp in complex with AcQLDLA | | Descriptor: | (ACE)QLDLA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3S
 
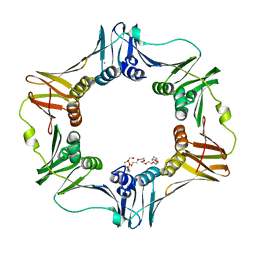 | | E. coli sliding clamp in P1 crystal space group | | Descriptor: | CALCIUM ION, DNA polymerase III subunit beta, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
7EL2
 
 | |
2WV0
 
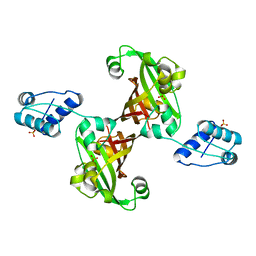 | | Crystal structure of the GntR-HutC family member YvoA from Bacillus subtilis | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REPRESSOR YVOA, SULFATE ION | | Authors: | Resch, M, Schiltz, E, Titgemeyer, F, Muller, Y.A. | | Deposit date: | 2009-10-12 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight Into the Induction Mechanism of the Gntr/Hutc Bacterial Transcription Regulator Yvoa
Nucleic Acids Res., 38, 2010
|
|
6ODM
 
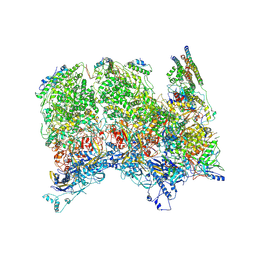 | | Herpes simplex virus type 1 (HSV-1) portal vertex-adjacent capsid/CATC, asymmetric unit | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
4K3O
 
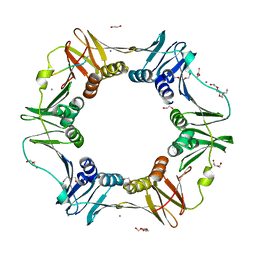 | | E. coli sliding clamp in complex with AcQADLF | | Descriptor: | (ACE)QADLF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
3KCC
 
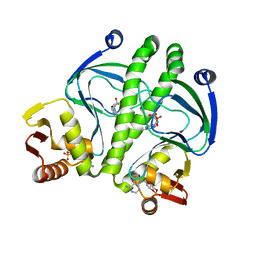 | | Crystal structure of D138L mutant of Catabolite Gene Activator Protein | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Tao, W.B, Gao, Z.Q, Zhou, J.H, Dong, Y.H, Yu, S.N. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The 1.6A resolution structure of activated D138L mutant of catabolite gene activator protein with two cAMP bound in each monomer
Int.J.Biol.Macromol., 48, 2011
|
|
4K3P
 
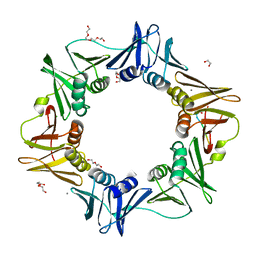 | | E. coli sliding clamp in complex with AcQLALF | | Descriptor: | (ACE)QLALF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
