2MOI
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MOL
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MPN
 
 | | 3D NMR structure of the transmembrane domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-05-29 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2N1F
 
 | | Structure and assembly of the mouse ASC filament by combined NMR spectroscopy and cryo-electron microscopy | | Descriptor: | Apoptosis-associated speck-like protein | | Authors: | Sborgi, L, Ravotti, F, Dandey, V, Dick, M, Mazur, A, Reckel, S, Chami, M, Scherer, S, Bockmann, A, Egelman, E, Stahlberg, H, Broz, P, Meier, B, Hiller, S. | | Deposit date: | 2015-04-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2JZZ
 
 | |
2M1J
 
 | | Ovine Doppel Signal peptide (1-30) | | Descriptor: | Prion-like protein doppel | | Authors: | Pimenta, J, Viegas, A, Sardinha, J, Santos, A, Cantante, C, Dias, F.M.V, Soares, R, Cabrita, E.J, Fontes, C.M.G.A, Prates, J.A.M, Pereira, R.M.L.N. | | Deposit date: | 2012-11-28 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and SRP54M predicted interaction of the N-terminal sequence (1-30) of the ovine Doppel protein.
Peptides, 49C, 2013
|
|
2KKE
 
 | | Solution NMR Structure of a dimeric protein of unknown function from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target TR5 | | Descriptor: | Uncharacterized protein | | Authors: | Swapna, G.V.T, Gunsalus, X, Huang, L, Xiao, K, Everett, J.K, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-18 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a putative uncharacterized protein from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target:TR5
To be Published
|
|
2K5B
 
 | | Human CDC37-HSP90 docking model based on NMR | | Descriptor: | Heat shock protein HSP 90-alpha, Hsp90 co-chaperone Cdc37 | | Authors: | Sreeramulu, S, Jonker, H.R.A, Lancaster, C.R, Richter, C, Langer, T, Schwalbe, H. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The human Cdc37.Hsp90 complex studied by heteronuclear NMR spectroscopy
J.Biol.Chem., 284, 2009
|
|
2KB5
 
 | | Solution NMR Structure of Eosinophil Cationic Protein/RNase 3 | | Descriptor: | Eosinophil cationic protein | | Authors: | Rico, M, Bruix, M, Laurents, D.V, Santoro, J, Jimenez, M, Boix, E, Moussaoui, M, Nogues, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-06-23 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The (1)H, (13)C, (15)N resonance assignment, solution structure, and residue level stability of eosinophil cationic protein/RNase 3 determined by NMR spectroscopy
Biopolymers, 91, 2009
|
|
2K88
 
 | | Association of subunit d (Vma6p) and E (Vma4p) with G (Vma10p) and the NMR solution structure of subunit G (G1-59) of the Saccharomyces cerevisiae V1VO ATPase | | Descriptor: | Vacuolar proton pump subunit G | | Authors: | Sankaranarayanan, N, Gayen, S, Thaker, Y, Subramanian, V, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Assembly of subunit d (Vma6p) and G (Vma10p) and the NMR solution structure of subunit G (G(1-59)) of the Saccharomyces cerevisiae V(1)V(O) ATPase.
Biochim.Biophys.Acta, 1787, 2009
|
|
2K8I
 
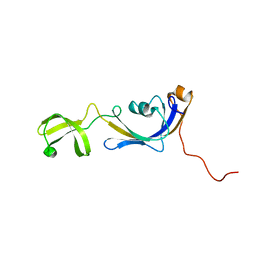 | | Solution structure of E.Coli SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Weininger, U, Balbach, J. | | Deposit date: | 2008-09-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of SlyD from Escherichia coli: spatial separation of prolyl isomerase and chaperone function.
J.Mol.Biol., 387, 2009
|
|
2K9C
 
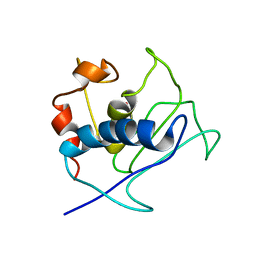 | | Paramagnetic shifts in solid-state NMR of Proteins to elicit structural information | | Descriptor: | COBALT (II) ION, Macrophage metalloelastase | | Authors: | Balayssac, S, Bertini, I, Bhaumik, A, Lelli, M, Luchinat, C. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic shifts in solid-state NMR of proteins to elicit structural information
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2M56
 
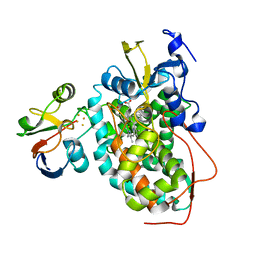 | | The structure of the complex of cytochrome P450cam and its electron donor putidaredoxin determined by paramagnetic NMR spectroscopy | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hiruma, Y, Hass, M.A.S, Ubbink, M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
J.Mol.Biol., 425, 2013
|
|
2K6I
 
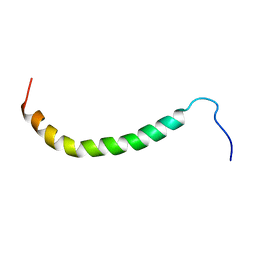 | | The domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H1-47 | | Descriptor: | Uncharacterized protein MJ0223 | | Authors: | Biukovic, N, Gayen, S, Pervushin, K, Gruber, G, Biukovic, G. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H(1-47).
Biophys.J., 97, 2009
|
|
2L03
 
 | |
2LHS
 
 | |
2LK7
 
 | |
2LU1
 
 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
2MJX
 
 | | Solution NMR structure of a mismatch DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*AP*CP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*CP*G)-3') | | Authors: | Ghosh, A, Kumar, K.R, Bhunia, A, Chatterjee, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Double GC:GC mismatch in dsDNA enhances local dynamics retaining the DNA footprint: a high-resolution NMR study
Chemmedchem, 9, 2014
|
|
2LXZ
 
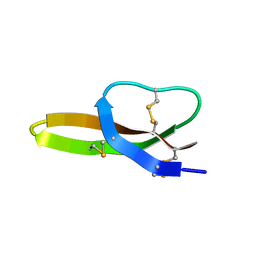 | | Solution Structure of the Antimicrobial Peptide Human Defensin 5 | | Descriptor: | Defensin-5 | | Authors: | Wommack, A.J, Robson, S.A, Wanniarahchi, Y.A, Wan, A, Turner, C.J, Nolan, E.M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and condition-dependent oligomerization of the antimicrobial Peptide human defensin 5.
Biochemistry, 51, 2012
|
|
2K3B
 
 | |
2JWQ
 
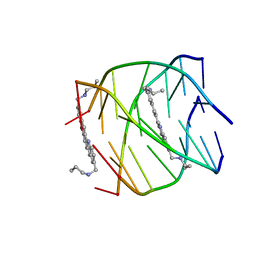 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
2N28
 
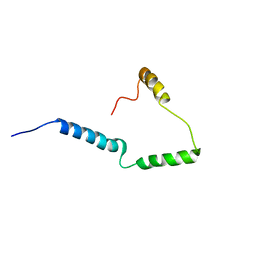 | | Solid-state NMR structure of Vpu | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2NPV
 
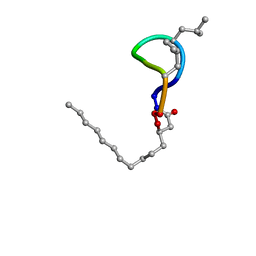 | |
2M9S
 
 | |
