6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
6M4K
 
 | | X-ray crystal structure of wild type alpha-amylase I from Eisenia fetida | | Descriptor: | ACETATE ION, Alpha-amylase, CALCIUM ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6LYH
 
 | | Crystal structure of tea N9-methyltransferase CkTcS in complex with SAH and 1,3,7-trimethyluric acid | | Descriptor: | 1,3,7-trimethyl-9H-purine-2,6,8-trione, N-methyltransferase CkTcS, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, Y, Zhang, Z.-M. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15000319 Å) | | Cite: | Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
Nat Commun, 11, 2020
|
|
6M08
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor. | | Descriptor: | (2S)-2-[(Z)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M4T
 
 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*(UD)P*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), N4-[4-[(6-chloranyl-2-methoxy-acridin-9-yl)amino]butyl]-1,3,5-triazine-2,4,6-triamine | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B, Wu, P.C. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
6M53
 
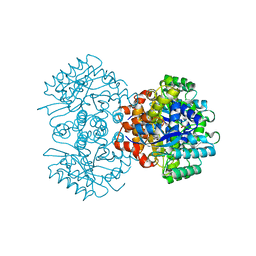 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, GLYCEROL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
6M5U
 
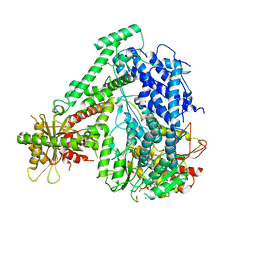 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M6B
 
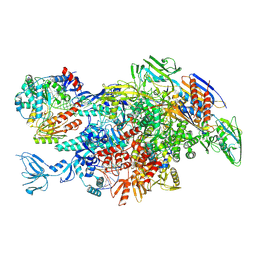 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M17
 
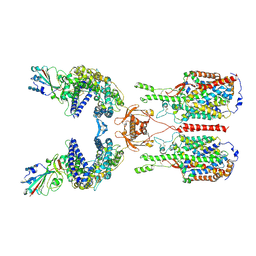 | | The 2019-nCoV RBD/ACE2-B0AT1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2020-02-24 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
6M2K
 
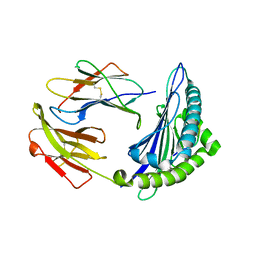 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-10 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M31
 
 | |
6M87
 
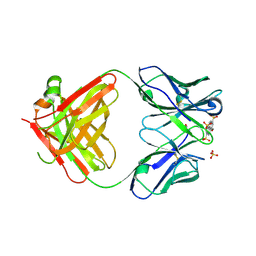 | | Fab 10A6 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, Fab 10A6 heavy chain, Fab 10A6 light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.
Biochemistry, 58, 2019
|
|
6M9A
 
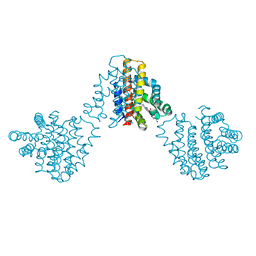 | | Bordetella pertussis globin coupled sensor regulatory domain (BpeGReg) | | Descriptor: | CALCIUM ION, HYDROGEN PEROXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rivera, S.R, Hoffer, E.D, Dunham, C.M, Weinert, E.E. | | Deposit date: | 2018-08-23 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into Oxygen-Dependent Signal Transduction within Globin Coupled Sensors.
Inorg Chem, 57, 2018
|
|
6M9S
 
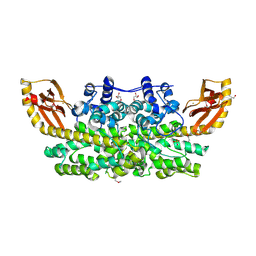 | | Crystal structure of SeMet SznF from Streptomyces achromogenes var. streptozoticus NRRL 2697 | | Descriptor: | FE (III) ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Rohac, R, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | An N-nitrosating metalloenzyme constructs the pharmacophore of streptozotocin.
Nature, 566, 2019
|
|
6M3Q
 
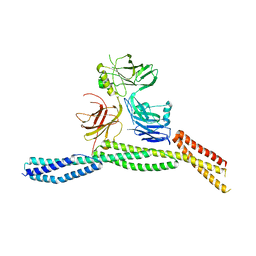 | | Crystal structure of AnkB/beta4-spectrin complex | | Descriptor: | Ankyrin-2, Spectrin beta chain | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.436 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
6M4C
 
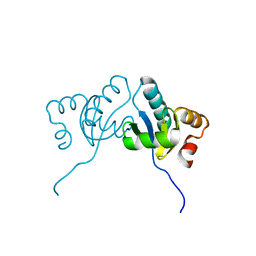 | | C. albicans actin interacting protein Aip5 | | Descriptor: | C. albicans actin interacting protein Aip5 | | Authors: | Loh, Z.Y, Gao, Y.G, Xie, Y, Miao, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Orchestrated actin nucleation by the Candida albicans polarisome complex enables filamentous growth.
J.Biol.Chem., 295, 2020
|
|
6M4E
 
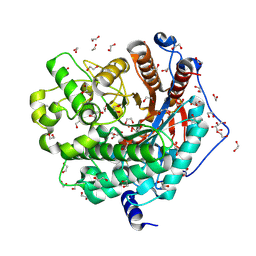 | | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M4Q
 
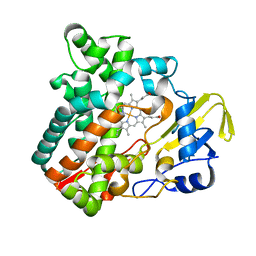 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
6M55
 
 | | Crystal structure of the E496A mutant of HsBglA in complex with 4-galactosyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-10 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M5J
 
 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA/TTCTGCAGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*TP*TP*CP*TP*GP*CP*AP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
6M5S
 
 | | The coordinates of the apo hexameric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6MB4
 
 | | Binary (sisomicin) structure of AAC-IIIb | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aac(3)-IIIb protein | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MBA
 
 | | Crystal Structure of Human Nav1.4 CTerminal Domain in Complex with apo Calmodulin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONATE ION, ... | | Authors: | Yoder, J, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Ca2+-dependent regulation of sodium channels NaV1.4 and NaV1.5 is controlled by the post-IQ motif.
Nat Commun, 10, 2019
|
|
6M67
 
 | | The Cryo-EM Structure of Human Pannexin 1 with D376E/D379E Mutation | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6M7F
 
 | | Wild-type Cucumene Synthase | | Descriptor: | Cucumene Synthase | | Authors: | Blank, P.N, Pemberton, T.A, Christianson, D.W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Crystal Structure of Cucumene Synthase, a Terpenoid Cyclase That Generates a Linear Triquinane Sesquiterpene.
Biochemistry, 57, 2018
|
|
