6LSU
 
 | | Crystal structure of Uso1-2 | | Descriptor: | Intracellular protein transport protein USO1 | | Authors: | Heo, Y.Y, Lee, H.H. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Uso1 membrane tether reveal an alternative conformation in the globular head domain
Sci Rep, 10, 2020
|
|
6LT5
 
 | | Lysozyme protected by alginate gel | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Tomoike, F, Morita, S, Nagae, T, Okada, T. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Post-crystallization protection of protein crystals
To Be Published
|
|
6LT6
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LUE
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with compound KL201 | | Descriptor: | 2-bromanyl-N-(5,6,7,8-tetrahydro-[1]benzothiolo[2,3-d]pyrimidin-4-yl)benzamide, Cryptochrome-1 | | Authors: | Miller, S, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Isoform-Selective Modulator of Cryptochrome 1 Regulates Circadian Rhythms in Mammals.
Cell Chem Biol, 27, 2020
|
|
6LUQ
 
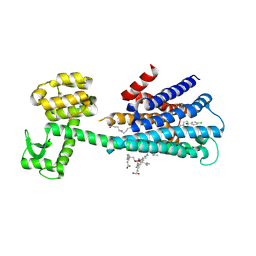 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
6LUT
 
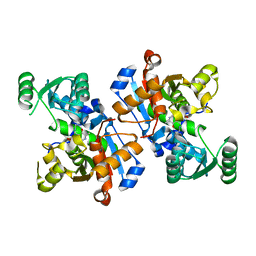 | |
6LT8
 
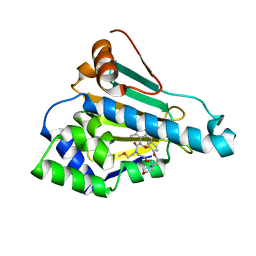 | | HSP90 in complex with KW-2478 | | Descriptor: | 2-[2-ethyl-6-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]carbonyl-3,5-bis(oxidanyl)phenyl]-~{N},~{N}-bis(2-methoxyethyl)ethanamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Anti-NSCLC activity in vitro of Hsp90 N inhibitor KW-2478 and complex crystal structure determination of Hsp90 N -KW-2478.
J.Struct.Biol., 213, 2021
|
|
6LT9
 
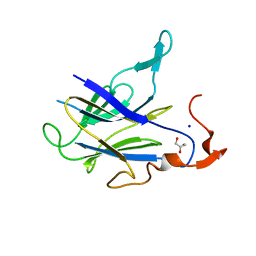 | |
6LUY
 
 | | Zn- Carbonic Anhydrase II pH 11.0 0 atm CO2 | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2020-02-02 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Elucidating the role of metal ions in carbonic anhydrase catalysis.
Nat Commun, 11, 2020
|
|
6LV5
 
 | | Ni- Carbonic Anhydrase II pH 7.8 0 atm CO2 | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, NICKEL (II) ION | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2020-02-02 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Elucidating the role of metal ions in carbonic anhydrase catalysis.
Nat Commun, 11, 2020
|
|
6LTH
 
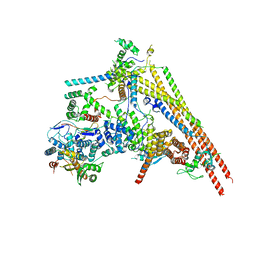 | | Structure of human BAF Base module | | Descriptor: | AT-rich interactive domain-containing protein 1A, SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
6LTY
 
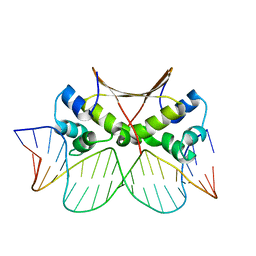 | | DNA bound antitoxin HigA3 | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*GP*AP*GP*AP*TP*AP*TP*AP*AP*CP*CP*TP*AP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*TP*AP*GP*GP*TP*TP*AP*TP*AP*TP*CP*TP*CP*GP*TP*GP*G)-3'), Putative antitoxin HigA3 | | Authors: | Park, J.Y, Lee, B.J. | | Deposit date: | 2020-01-23 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Induced DNA bending by unique dimerization of HigA antitoxin.
Iucrj, 7, 2020
|
|
6LVE
 
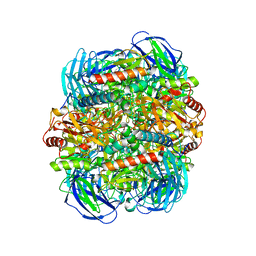 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LW3
 
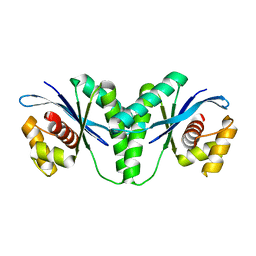 | | Crystal structure of RuvC from Pseudomonas aeruginosa | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC | | Authors: | Hu, Y, He, Y, Lin, Z. | | Deposit date: | 2020-02-07 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Biochemical and structural characterization of the Holliday junction resolvase RuvC from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LX2
 
 | | Potato D-enzyme complexed with CA26 | | Descriptor: | 4-alpha-glucanotransferase, chloroplastic/amyloplastic, 4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
6LX9
 
 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LTZ
 
 | |
6LU8
 
 | | Cryo-EM structure of a human pre-60S ribosomal subunit - state A | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-26 | | Release date: | 2020-08-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LXK
 
 | | Crystal structure of Z2B3 D102R Fab in complex with influenza virus neuraminidase from A/Serbia/NS-601/2014 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3-D102R Fab, Light chain of Z2B3-D102R Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.608 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LZG
 
 | | Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Wang, Q.H, Song, H, Qi, J.X. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2.
Cell, 181, 2020
|
|
6M0D
 
 | | Beijerinckia indica beta-fructosyltransferase | | Descriptor: | Levansucrase, MAGNESIUM ION | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6M0S
 
 | | 3.6A Yeast Vo state3 prime | | Descriptor: | Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Roh, S.H, Shekhar, M, Pintilie, G, Chipot, C, Wilkens, S, SIngharoy, A, Chiu, W. | | Deposit date: | 2020-02-22 | | Release date: | 2020-11-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Sci Adv, 6, 2020
|
|
6M2H
 
 | | A reaction intermediate of sirohydrochlorin nickelochelatase CfbA in complex with Ni2+ and sirohydrochlorin | | Descriptor: | 3,3',3'',3'''-[(7S,8S,12S,13S)-3,8,13,17-tetrakis(carboxymethyl)-8,13-dimethyl-7,8,12,13-tetrahydroporphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, NICKEL (II) ION, Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
6M2U
 
 | |
6M3T
 
 | |
