7NHM
 
 | | 70S ribosome from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Murina, V, Hauryliuk, V, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
7NFX
 
 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | Deposit date: | 2021-02-08 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
6HTQ
 
 | |
5MDY
 
 | | Structure of ArfA and TtRF2 bound to the 70S ribosome (pre-accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
6FXC
 
 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-21 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
5MLC
 
 | | Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions | | Descriptor: | 23S ribosomal RNA, chloroplastic, 4.8S ribosomal RNA, ... | | Authors: | Graf, M, Arenz, S, Huter, P, Doenhoefer, A, Novacek, J, Wilson, D.N. | | Deposit date: | 2016-12-06 | | Release date: | 2016-12-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions.
Nucleic Acids Res., 45, 2017
|
|
6TC3
 
 | |
6TBV
 
 | |
5MRF
 
 | | Structure of the yeast mitochondrial ribosome - Class C | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.97 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
6GC0
 
 | | 50S ribosomal subunit assembly intermediate state 4 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6UO1
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA (containing pseudouridine at the first position of the codon) and deacylated A-, P-, and E-site tRNAs at 2.95A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Dobosz-Bartoszek, M, Polikanov, Y.S. | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Pseudouridinylation of mRNA coding sequences alters translation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HHQ
 
 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.10000038 Å) | | Cite: | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
6T7I
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CGA inhibitory codon combination. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
7N30
 
 | | Elongating 70S ribosome complex in a hybrid-H2* pre-translocation (PRE-H2*) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-30 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2C
 
 | | Elongating 70S ribosome complex in a fusidic acid-stalled intermediate state of translocation bound to EF-G(GDP) (INT2) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N31
 
 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
6T59
 
 | | Structure of rabbit 80S ribosome translating beta-tubulin in complex with tetratricopeptide protein 5 and nascent chain-associated complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Lin, Z, Gasic, I, Chandrasekaran, V, Peters, N, Shao, S, Ramakrishnan, V, Mitchison, T.J, Hegde, R.S. | | Deposit date: | 2019-10-15 | | Release date: | 2019-11-27 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | TTC5 mediates autoregulation of tubulin via mRNA degradation.
Science, 367, 2020
|
|
5NGM
 
 | | 2.9S structure of the 70S ribosome composing the S. aureus 100S complex | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Amunts, A, Yonath, A. | | Deposit date: | 2017-03-18 | | Release date: | 2017-10-04 | | Last modified: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
5NJT
 
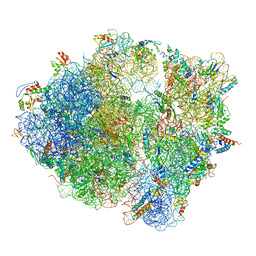 | | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Beckert, B, Abdelshahid, M, Schaefer, H, Steinchen, W, Arenz, S, Berninghausen, O, Beckmann, R, Bange, G, Turgay, K, Wilson, D.N. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization.
EMBO J., 36, 2017
|
|
6I1P
 
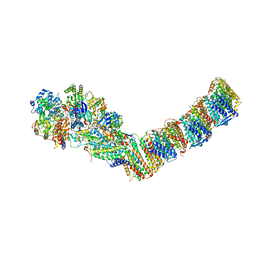 | | Respiratory complex I from Thermus thermophilus with bound NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-10-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
5MMM
 
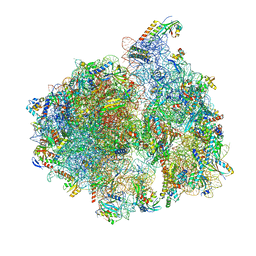 | | Structure of the 70S chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein 2, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
7N2U
 
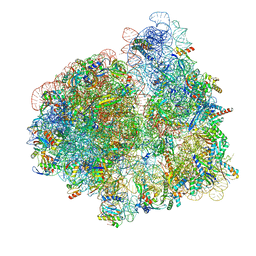 | | Elongating 70S ribosome complex in a hybrid-H1 pre-translocation (PRE-H1) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N1P
 
 | | Elongating 70S ribosome complex in a classical pre-translocation (PRE-C) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, S.K, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
6HCJ
 
 | | Structure of the rabbit 80S ribosome on globin mRNA in the rotated state with A/P and P/E tRNAs | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCQ
 
 | | Structure of the rabbit collided di-ribosome (collided monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
