8FDQ
 
 | |
6CE6
 
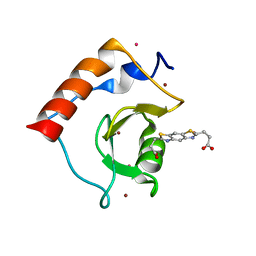 | | Structure of HDAC6 zinc-finger ubiquitin binding domain soaked with 3,3'-(benzo[1,2-d:5,4-d']bis(thiazole)-2,6-diyl)dipropionic acid | | Descriptor: | 3,3'-(benzo[1,2-d:5,4-d']bis[1,3]thiazole-2,6-diyl)dipropanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CED
 
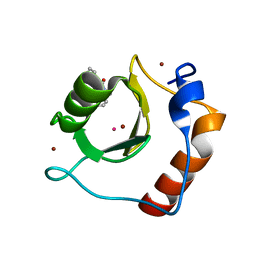 | | Crystal structure of fragment 3-(3-Methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6DF4
 
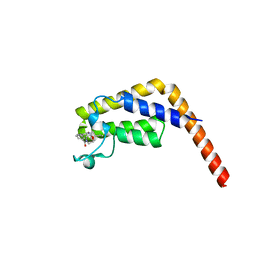 | | TAF1-BD2 in complex with Cpd8 (6-(but-3-en-1-yl)-4-(3-(morpholine-4-carbonyl)phenyl)-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one) | | Descriptor: | 6-(but-3-en-1-yl)-4-[3-(morpholine-4-carbonyl)phenyl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Transcription initiation factor TFIID subunit | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2018-05-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | GNE-371, a Potent and Selective Chemical Probe for the Second Bromodomains of Human Transcription-Initiation-Factor TFIID Subunit 1 and Transcription-Initiation-Factor TFIID Subunit 1-like.
J. Med. Chem., 61, 2018
|
|
6DF7
 
 | |
6GVF
 
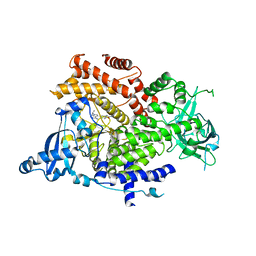 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidin-4-ylamine | | Descriptor: | 5-(4-azanyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6D88
 
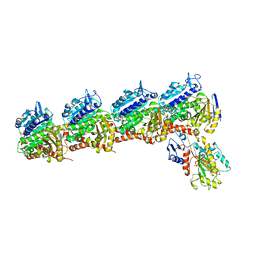 | | Tubulin-RB3_SLD-TTL in complex with compound 13f | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2018-04-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural Modification of the 3,4,5-Trimethoxyphenyl Moiety in the Tubulin Inhibitor VERU-111 Leads to Improved Antiproliferative Activities.
J. Med. Chem., 61, 2018
|
|
6NVJ
 
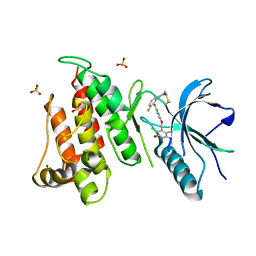 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
5KYK
 
 | | Covalent GTP-competitive inhibitors of KRAS G12C: Guanosine bisphosphonate Analogs | | Descriptor: | 5'-O-[(R)-[({2-[(chloroacetyl)amino]ethyl}sulfamoyl)methyl](hydroxy)phosphoryl]guanosine, GTPase KRas | | Authors: | Xiong, Y, Lu, J, Hunter, J, Li, L, Scott, D, Manandhar, A, Gondi, S, Westover, K.D, Gray, N.S. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Covalent Guanosine Mimetic Inhibitors of G12C KRAS.
ACS Med Chem Lett, 8, 2017
|
|
6BS2
 
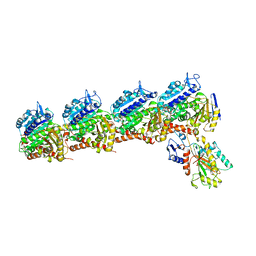 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 8b | | Descriptor: | 1-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6NVG
 
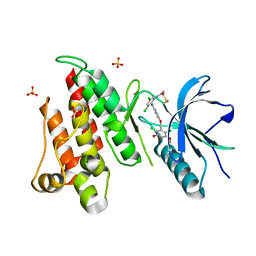 | | FGFR4 complex with N-(3,5-dichloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)phenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3,5-dichloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)phenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVK
 
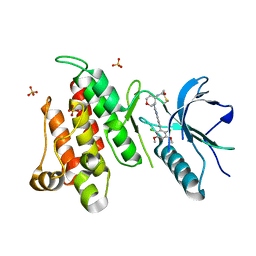 | | FGFR4 complex with BLU-554, N-((3S,4S)-3-((6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl)amino)tetrahydro-2H-pyran-4-yl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3S,4S)-3-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl]amino}oxan-4-yl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6CEA
 
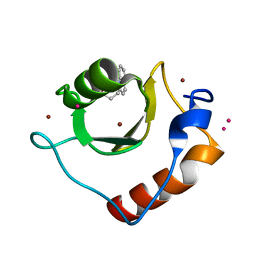 | | Crystal structure of fragment 3-(quinolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(quinolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEE
 
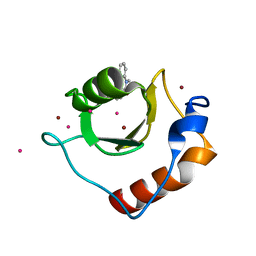 | | Crystal structure of fragment 3-(1-Methyl-2-oxo-1,2-dihydroquinoxalin-3-yl)propionic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(4-methyl-3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6NVI
 
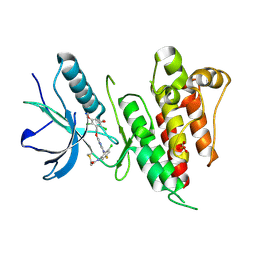 | | FGFR4 complex with N-(3-chloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-5-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3-chloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-5-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
5M5J
 
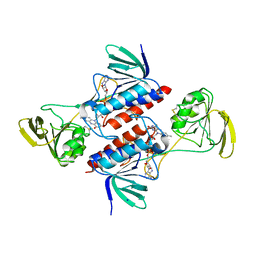 | | Thioredoxin reductase from Giardia duodenalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, Thioredoxin reductase | | Authors: | Fiorillo, A, Ilari, A, Lalle, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural characterization of Giardia duodenalis thioredoxin reductase (gTrxR) and computational analysis of its interaction with NBDHEX.
Eur J Med Chem, 135, 2017
|
|
6G5J
 
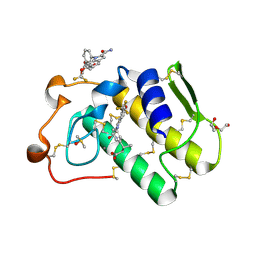 | | Secreted phospholipase A2 type X in complex with ligand | | Descriptor: | (3~{R})-3-[3-[2-aminocarbonyl-6-(trifluoromethyloxy)indol-1-yl]phenyl]butanoic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Selective sPLA2-X Inhibitor (-)-2-{2-[Carbamoyl-6-(trifluoromethoxy)-1H-indol-1-yl]pyridine-2-yl}propanoic Acid.
ACS Med Chem Lett, 9, 2018
|
|
6BRY
 
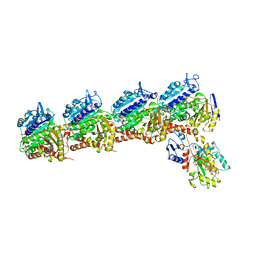 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 6a | | Descriptor: | 1-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
7TBA
 
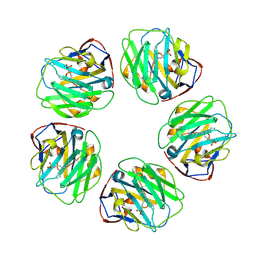 | | Pentraxin - ligand complex | | Descriptor: | C-reactive protein, CALCIUM ION, [3-(dibutylamino)propyl]phosphonic acid | | Authors: | Shing, K.S.C.T, Morton, C.J, Parker, M.W. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A novel phosphocholine-mimetic inhibits a pro-inflammatory conformational change in C-reactive protein.
Embo Mol Med, 15, 2023
|
|
6NVH
 
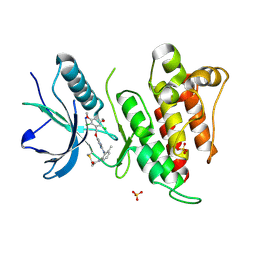 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6DGT
 
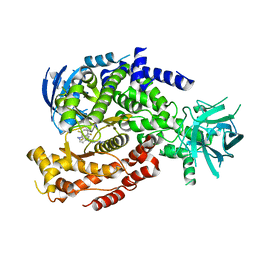 | | Selective PI3K beta inhibitor bound to PI3K delta | | Descriptor: | 4-[1-(5,8-difluoroquinolin-4-yl)-2-methyl-4-(4H-1,2,4-triazol-3-yl)-1H-benzimidazol-6-yl]-3-fluoropyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J, Villasenor, A, McGrath, M. | | Deposit date: | 2018-05-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Atropisomerism by Design: Discovery of a Selective and Stable Phosphoinositide 3-Kinase (PI3K) beta Inhibitor.
J. Med. Chem., 61, 2018
|
|
6SZJ
 
 | |
6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6BRF
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[3,2-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
