7TW8
 
 | |
6NHB
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(5-(2-((2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(5-{2-[(2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl]ethyl}-2,3-difluorophenyl)ethyl]-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
7Q3I
 
 | |
7NMO
 
 | | Crystal structure of beta-2-microglobulin D76A mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5HS1
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) complexed with Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Sabherwal, M, Sagatova, A, Keniya, M.V, Wilson, R.K, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2016-01-24 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
7TW7
 
 | |
5E9V
 
 | | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(imidazo[1,2-a]pyridin-5-yl)-7-(morpholin-4-yl)indolizin-3-yl]ethanone, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Hay, D.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Schofield, C.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand
To Be Published
|
|
5HLB
 
 | | E. coli PBP1b in complex with acyl-aztreonam and moenomycin | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Escherichia coli Penicillin-Binding Protein 1B: Structural Insights into Inhibition.
J. Biol. Chem., 2016
|
|
5EA1
 
 | | Crystal Structure of SMARCA4 bromodomain in complex with MPD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Transcription activator BRG1 | | Authors: | Lolli, G, Caflisch, A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Throughput Fragment Docking into the BAZ2B Bromodomain: Efficient in Silico Screening for X-Ray Crystallography.
Acs Chem.Biol., 11, 2016
|
|
7NMR
 
 | | Crystal structure of beta-2-microglobulin D76S mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMT
 
 | | Crystal structure of beta-2-microglobulin D76G mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8SKQ
 
 | |
6NPS
 
 | | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus | | Descriptor: | AxyAgu115A, CHLORIDE ION, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yan, R, Master, E, Savchenko, A. | | Deposit date: | 2019-01-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural characterization of the family GH115 alpha-glucuronidase from Amphibacillus xylanus yields insight into its coordinated action with alpha-arabinofuranosidases.
N Biotechnol, 2021
|
|
7Q14
 
 | |
6NQM
 
 | | Crystal structure of Human LSD1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A | | Authors: | Tan, A.H.Y, Tu, W, McCuaig, R, Donovan, T, Tsimbalyuk, S, Forwood, J.K, Rao, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lysine-Specific Histone Demethylase 1A Regulates Macrophage Polarization and Checkpoint Molecules in the Tumor Microenvironment of Triple-Negative Breast Cancer.
Front Immunol, 10, 2019
|
|
5EA8
 
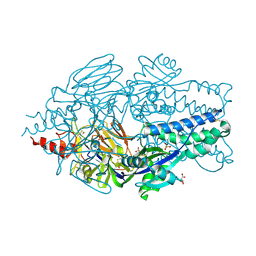 | | Crystal Structure of Prefusion RSV F Glycoprotein Fusion Inhibitor Resistance Mutant D489Y | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, D(-)-TARTARIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
7NMC
 
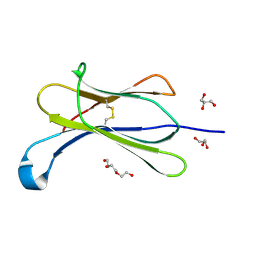 | | Crystal structure of beta-2-microglobulin D76E mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8VRV
 
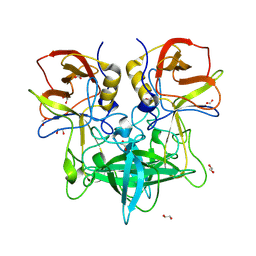 | | GII.4c H2-tri HBGA norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, FORMIC ACID, ... | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2024-01-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Advancement of norovirus therapies
To Be Published
|
|
7NMV
 
 | | Crystal structure of beta-2-microglobulin D76Q mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZN6
 
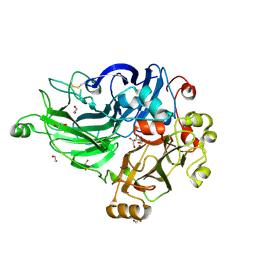 | | Crystal structure of laccase-like multicopper oxidase (LMCO) from Thermothelomyces thermophilus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kosinas, C, Dimarogona, M, Zerva, A, Topakas, E. | | Deposit date: | 2022-04-20 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function studies of a novel laccase-like multicopper oxidase from Thermothelomyces thermophila provide insights into its biological role.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5V8I
 
 | | Thermus thermophilus 70S ribosome lacking ribosomal protein uS17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Gregory, S.T, Jogl, G. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural robustness of the ribosome inferred from X-ray crystal structures of the 30S ribosomal subunit and the 70S ribosome lacking ribosomal protein uS17
To be published
|
|
8SY3
 
 | |
7TGM
 
 | |
5V31
 
 | | Ethylene forming enzyme in complex with manganese and L-arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
7NNK
 
 | | Crystal structure of S116A mutant of hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-02-25 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
