4FMH
 
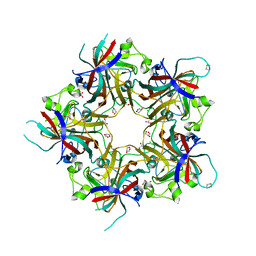 | | Merkel Cell Polyomavirus VP1 in complex with Disialyllactose | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4FOE
 
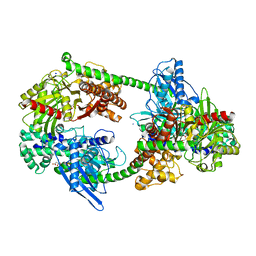 | |
4K96
 
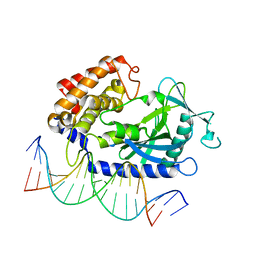 | | Structure of Binary Complex of cGAS with Bound dsDNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA-F, DNA-R, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
3UOY
 
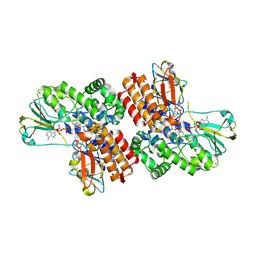 | | Crystal Structure of OTEMO complex with FAD and NADP (form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO, ... | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3RIM
 
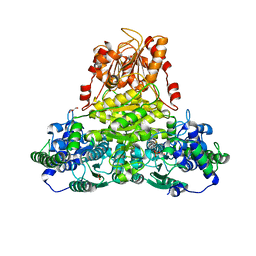 | | Crystal structure of mycobacterium tuberculosis Transketolase (Rv1449c) | | Descriptor: | GLYCEROL, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Pojer, F, Fullam, E, Jones, T.A, Cole, S.T. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and function of the transketolase from Mycobacterium tuberculosis and comparison with the human enzyme.
Open Biol, 2, 2012
|
|
4HBW
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-ethyl-3-methyl-2-oxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4K6W
 
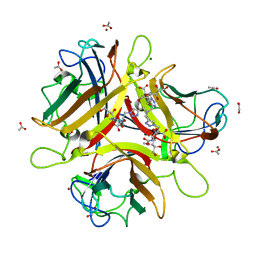 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0408 | | Descriptor: | 1,2-ETHANEDIOL, 2,2',2''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]triethanol, 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
3UU3
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4HCU
 
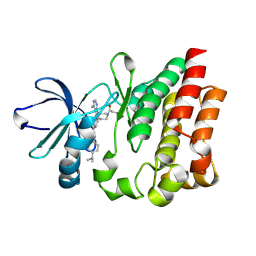 | | Crystal structure of ITK in complext with compound 40 | | Descriptor: | 3-{4-amino-1-[(3R)-1-propanoylpiperidin-3-yl]-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-[4-(propan-2-yl)phenyl]benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
4KGC
 
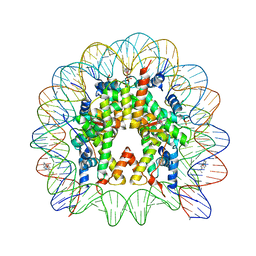 | | Nucleosome Core Particle Containing (ETA6-P-CYMENE)-(1, 2-ETHYLENEDIAMINE)-RUTHENIUM | | Descriptor: | (ethane-1,2-diamine-kappa~2~N,N')[(1,2,3,4,5,6-eta)-1-methyl-4-(propan-2-yl)cyclohexane-1,2,3,4,5,6-hexayl]ruthenium, DNA (145-mer), Histone H2A, ... | | Authors: | Adhireksan, Z, Davey, C.A. | | Deposit date: | 2013-04-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Ligand substitutions between ruthenium-cymene compounds can control protein versus DNA targeting and anticancer activity
Nat Commun, 5, 2014
|
|
3V0U
 
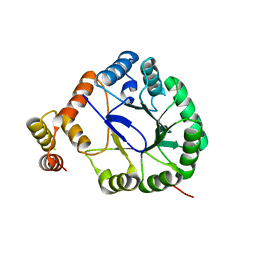 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3UOV
 
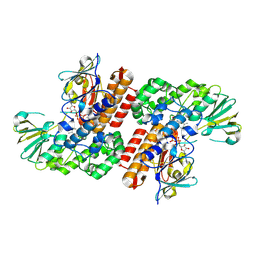 | | Crystal Structure of OTEMO (FAD bound form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP5
 
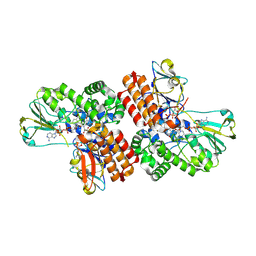 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
4JAR
 
 | |
3QAS
 
 | | Structure of Undecaprenyl Diphosphate synthase | | Descriptor: | Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Oldfield, E. | | Deposit date: | 2011-01-11 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Applying Molecular Dynamics Simulations to Identify Rarely Sampled Ligand-bound Conformational States of Undecaprenyl Pyrophosphate Synthase, an Antibacterial Target.
Chem.Biol.Drug Des., 77, 2011
|
|
4JAO
 
 | |
4GX9
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
4FMG
 
 | |
4LCC
 
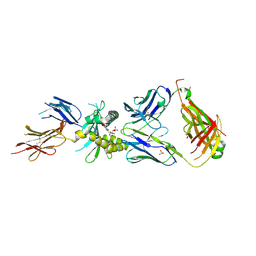 | | Crystal structure of a human MAIT TCR in complex with a bacterial antigen bound to humanized bovine MR1 | | Descriptor: | 1-deoxy-1-[6-(hydroxymethyl)-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl]-D-arabinitol, Beta-2-microglobulin, MHC class I-related protein, ... | | Authors: | Lopez-Sagaseta, J, Adams, E.J. | | Deposit date: | 2013-06-21 | | Release date: | 2013-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | MAIT Recognition of a Stimulatory Bacterial Antigen Bound to MR1.
J.Immunol., 191, 2013
|
|
3RPI
 
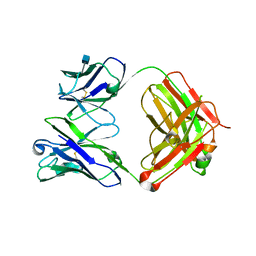 | | Crystal Structure of Fab from 3BNC60, Highly Potent anti-HIV Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from highly potent anti-HIV neutralizing antibody, Light chain from highly potent anti-HIV neutralizing antibody | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding.
Science, 333, 2011
|
|
3UP4
 
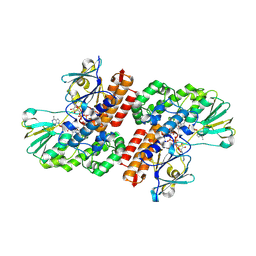 | | Crystal Structure of OTEMO complex with FAD and NADP (form 3) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
4LKE
 
 | | Crystal Structure of Pseudomonas aeruginosa Lectin LecA Complexed with GalA-WRI at 1.65 A Resolution | | Descriptor: | CALCIUM ION, P-HYDROXYBENZOIC ACID, PA-I galactophilic lectin, ... | | Authors: | Kadam, R.U, Stocker, A, Reymond, J.-L. | | Deposit date: | 2013-07-07 | | Release date: | 2013-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Structure-Based Optimization of the Terminal Tripeptide in Glycopeptide Dendrimer Inhibitors of Pseudomonas aeruginosa Biofilms Targeting LecA.
Chemistry, 19, 2013
|
|
3UU5
 
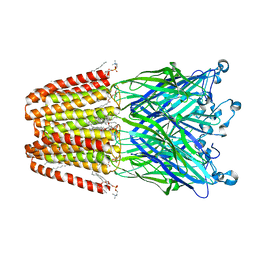 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0S
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3UU8
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
