3VK6
 
 | |
3PG6
 
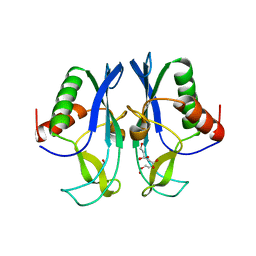 | | The carboxyl terminal domain of human deltex 3-like | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase DTX3L, ... | | Authors: | Walker, J.R, Obiero, J, Kania, J, Schuler, H, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-30 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fold of the conserved DTC domain in Deltex proteins.
Proteins, 80, 2012
|
|
1AKU
 
 | | D95A HYDROQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1AKQ
 
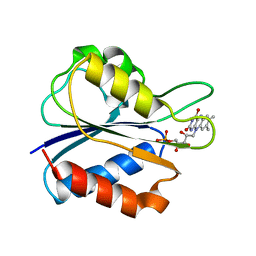 | | D95A OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-03-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1AKV
 
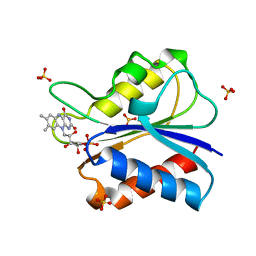 | | D95A SEMIQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
8EQM
 
 | | Structure of a dimeric photosystem II complex acclimated to far-red light | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Shen, G, Flesher, D.A, Kurashov, V, Golbeck, J.H, Brudvig, G.W, Amin, M, Bryant, D.A. | | Deposit date: | 2022-10-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a dimeric photosystem II complex from a cyanobacterium acclimated to far-red light.
J.Biol.Chem., 299, 2022
|
|
5F5L
 
 | |
1NBE
 
 | |
7PUI
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 2-pyridineboronic acid | | Descriptor: | Cholinephosphate cytidylyltransferase, pyridin-2-ylboronic acid | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7PVE
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 4-bromo-1H-imidazole | | Descriptor: | 4-bromo-1H-imidazole, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7PVG
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with isonicotinic acid | | Descriptor: | Cholinephosphate cytidylyltransferase, pyridine-4-carboxylic acid | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
8CKF
 
 | | Crystal Structure of the first bromodomain of human BRD4 L94C variant in complex with racemic 3,5-dimethylisoxazol ligand | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{S})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Thomas, A.M, McDonough, M.A, Schiedel, M, Conway, S.J. | | Deposit date: | 2023-02-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutate and Conjugate: A Method to Enable Rapid In-Cell Target Validation.
Acs Chem.Biol., 18, 2023
|
|
7PVF
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 3-hydroxy-1lambda6-thietane-1,1-dione | | Descriptor: | 1,1-bis(oxidanylidene)thietan-3-ol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
8EUQ
 
 | | Crystal structure of HLA-DRA*01:01/HLA-DRB1*04:01 in complex with c44H10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, ... | | Authors: | Kassardjian, A, Julien, J.-P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Modular adjuvant-free pan-HLA-DR-immunotargeting subunit vaccine against SARS-CoV-2 elicits broad sarbecovirus-neutralizing antibody responses.
Cell Rep, 42, 2023
|
|
7Q3M
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 3-hydroxypiperidine | | Descriptor: | (3S)-piperidin-3-ol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7Q2V
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1-methylpyrrolidin-3-ol | | Descriptor: | (3R)-1-methylpyrrolidin-3-ol, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q2K
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 2-pyrrolidinone | | Descriptor: | Cholinephosphate cytidylyltransferase, pyrrolidin-2-one | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7Q3W
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with (R)-2-Aminobutanamide hydrochloride | | Descriptor: | (R)-2-Aminobutanamide, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7Q2I
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Tetrahydrofurfurylamine | | Descriptor: | 1-[(2R)-oxolan-2-yl]methanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q2L
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 3-Aminopyrrolidin-2-one hydrochloride | | Descriptor: | (3R)-3-aminopyrrolidin-2-one, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q2M
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with but-3-yn-2-amine hydrochloride | | Descriptor: | (2S)-but-3-yn-2-amine, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
1X84
 
 | | IPP isomerase (wt) reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
7PUX
 
 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
8EXI
 
 | |
8EXJ
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain in complex with a JAK1 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK1 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
