2I4Z
 
 | |
6KYQ
 
 | |
6KX5
 
 | |
2HZQ
 
 | |
6KXF
 
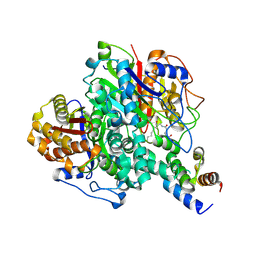 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6KXR
 
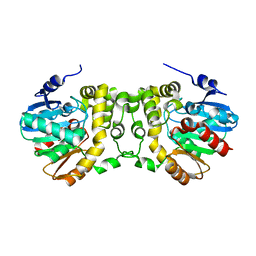 | | Crystal structure of wild type Alp1U from the biosynthesis of kinamycins | | Descriptor: | D-MALATE, Putative hydrolase | | Authors: | Zhang, L, Yingli, Z, De, B.C, Zhang, C. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45073676 Å) | | Cite: | Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha / beta-fold epoxide hydrolase Alp1U.
J.Biol.Chem., 295, 2020
|
|
6KZ0
 
 | | HRV14 3C in complex with single chain antibody GGVV | | Descriptor: | GGVV H chain, GGVV L chain, Genome polyprotein | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KZM
 
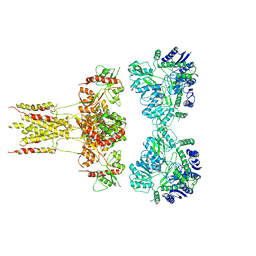 | | GluK3 receptor complex with kainate | | Descriptor: | Glutamate receptor ionotropic, kainate 3 | | Authors: | Kumar, J, Kumari, J, Burada, A.P. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural dynamics of the GluK3-kainate receptor neurotransmitter binding domains revealed by cryo-EM.
Int.J.Biol.Macromol., 149, 2020
|
|
6KY0
 
 | |
6KY8
 
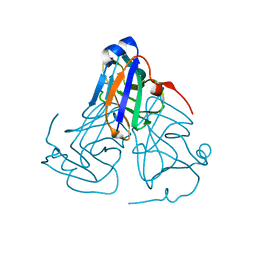 | | Crystal structure of ASFV dUTPase | | Descriptor: | E165R | | Authors: | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
6KYC
 
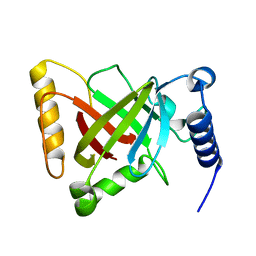 | | Structure of the S207A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
6KYE
 
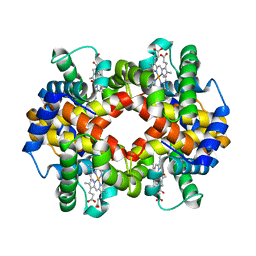 | | The crystal structure of recombinant human adult hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Kihira, K, Funaki, R, Okamoto, W, Endo, C, Morita, Y, Komatsu, T. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Genetically engineered haemoglobin wrapped covalently with human serum albumins as an artificial O2carrier.
J Mater Chem B, 8, 2020
|
|
2I1W
 
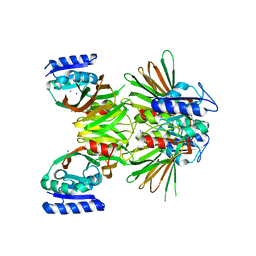 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes | | Descriptor: | IODIDE ION, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | Deposit date: | 2006-08-15 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
6L0T
 
 | |
6L0Y
 
 | | Structure of dsRNA with G-U wobble base pairs | | Descriptor: | RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3') | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of dsRNA with G-U wobble base pairs
To Be Published
|
|
6L16
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6KYV
 
 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
2I4P
 
 | |
6L1I
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
2I51
 
 | |
6L21
 
 | | Crystal structure of CK2a1 H160A with hematein | | Descriptor: | (6aR)-3,4,6a,10-tetrakis(oxidanyl)-6,7-dihydroindeno[2,1-c]chromen-9-one, Casein kinase II subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05451775 Å) | | Cite: | Structural insights for producing CK2 alpha 1-specific inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2I5T
 
 | | Crystal Structure of hypothetical protein LOC79017 from Homo sapiens | | Descriptor: | Protein C7orf24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Homo sapiens protein LOC79017.
Proteins, 70, 2008
|
|
6L2C
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase in complex with CoA | | Descriptor: | Acetyl-CoA-acetyltransferase, putative, COENZYME A | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
6L1R
 
 | |
