2KYV
 
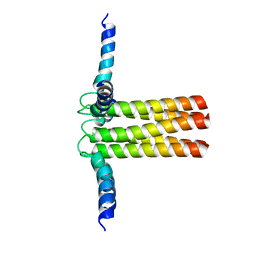 | | Hybrid solution and solid-state NMR structure ensemble of phospholamban pentamer | | Descriptor: | Phospholamban | | Authors: | Verardi, R, Shi, L, Traaseth, N.J, Veglia, G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2KUM
 
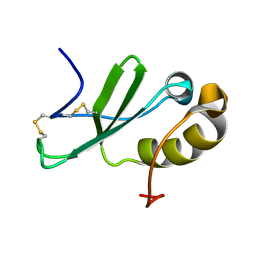 | | Solution structure of the human chemokine CCL27 | | Descriptor: | C-C motif chemokine 27 | | Authors: | Kirkpatrick, J.P, Jansma, A, Hsu, A, Handel, T.M, Nietlispach, D. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27.
J.Biol.Chem., 285, 2010
|
|
2KXD
 
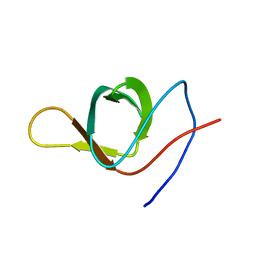 | | The structure of SH3-F2 | | Descriptor: | 11-mer peptide,Spectrin alpha chain, non-erythrocytic 1,Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Kutyshenko, V.P, Gushchina, L.V, Khristoforov, V.S, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, I.u.A, Fedyukina, D.V, Filimonov, V.V. | | Deposit date: | 2010-04-30 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the chimeric protein SH3-F2
Mol.Biol.(Engl.Transl.), 44, 2010
|
|
2KZN
 
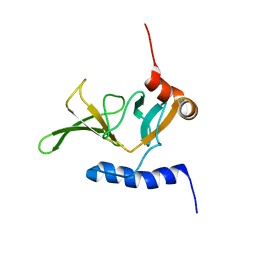 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1W0B
 
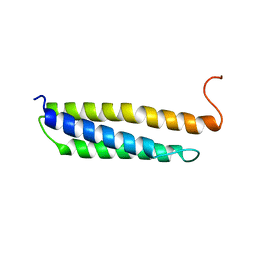 | | Solution structure of the human alpha-hemoglobin stabilizing protein (AHSP) P30A mutant | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1W0A
 
 | | Solution structure of the trans form of the human alpha-hemoglobin stabilizing protein (AHSP) | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1W09
 
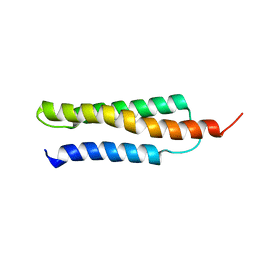 | | Solution structure of the cis form of the human alpha-hemoglobin stabilizing protein (AHSP) | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
1WQE
 
 | | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx3 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-28 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
2JYZ
 
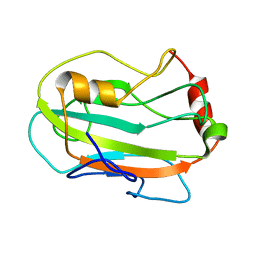 | | CG7054 solution structure | | Descriptor: | CG7054-PA | | Authors: | Rautureau, G, Jouvensal, L, Vovelle, F, Schoentgen, F, Locker, D, Decoville, M, Damblon, C. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a phosphatidyl-ethanolamine binding protein from Drosophila
Proteins, 78, 2009
|
|
1WQC
 
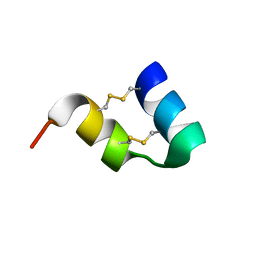 | | An unusual fold for potassium channel blockers : NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx1 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-27 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
2KW5
 
 | | Solution NMR Structure of the Slr1183 protein from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Rossi, P, Forouhar, F, Lee, H, Lange, O, Mao, B, Lemak, A, Maglaqui, M, Belote, R, Ciccosanti, C, Foote, E, Sahdev, S, Acton, T, Xiao, R, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LNU
 
 | | Solution NMR Structure of the uncharacterized protein from gene locus rrnAC0354 of Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR11 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Liu, G, Lange, O.F, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LK7
 
 | |
2LU1
 
 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
2M9R
 
 | |
2M9S
 
 | |
2JZ7
 
 | |
2MJX
 
 | | Solution NMR structure of a mismatch DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*AP*CP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*CP*G)-3') | | Authors: | Ghosh, A, Kumar, K.R, Bhunia, A, Chatterjee, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Double GC:GC mismatch in dsDNA enhances local dynamics retaining the DNA footprint: a high-resolution NMR study
Chemmedchem, 9, 2014
|
|
2K1G
 
 | | Solution NMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 | | Descriptor: | Lipoprotein spr | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad.
Biochemistry, 47, 2008
|
|
2LXZ
 
 | | Solution Structure of the Antimicrobial Peptide Human Defensin 5 | | Descriptor: | Defensin-5 | | Authors: | Wommack, A.J, Robson, S.A, Wanniarahchi, Y.A, Wan, A, Turner, C.J, Nolan, E.M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and condition-dependent oligomerization of the antimicrobial Peptide human defensin 5.
Biochemistry, 51, 2012
|
|
2N28
 
 | | Solid-state NMR structure of Vpu | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2N29
 
 | | Solution-state NMR structure of Vpu cytoplasmic domain | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2MOP
 
 | | Structure of Bitistatin A | | Descriptor: | Disintegrin bitistatin | | Authors: | Carbajo, R.J, Calvete, J, Sanz, L, Perez, A. | | Deposit date: | 2014-04-29 | | Release date: | 2014-11-12 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins.
Febs J., 282, 2015
|
|
2NPV
 
 | |
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | Descriptor: | Cold shock protein CspA | | Authors: | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
