7EWP
 
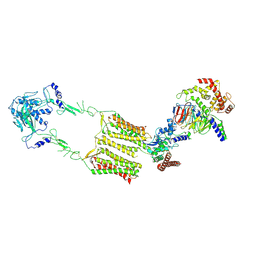 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:1:1 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7EWL
 
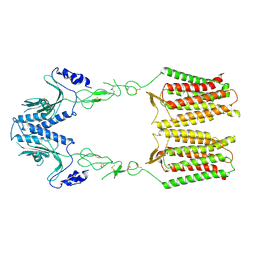 | | cryo-EM structure of apo GPR158 | | Descriptor: | Probable G-protein coupled receptor 158 | | Authors: | Jeong, E, Kim, Y, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7EWR
 
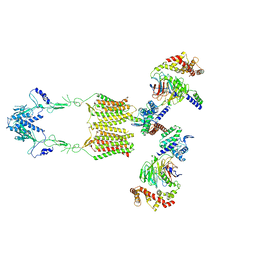 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:2:2 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
6PIZ
 
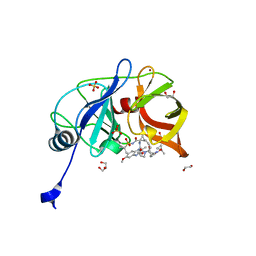 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-1 (NR02-24) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopropyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, GLYCEROL, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6P5K
 
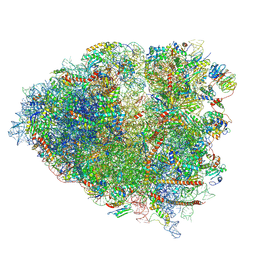 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
7EU0
 
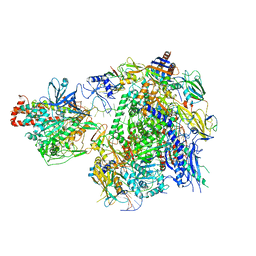 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 backtracked complex | | Descriptor: | DNA (33-MER), DNA (5'-D(*CP*TP*GP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*GP*AP*GP*TP*G)-3'), DNA-directed RNA polymerase IV subunit 1, ... | | Authors: | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
8AUP
 
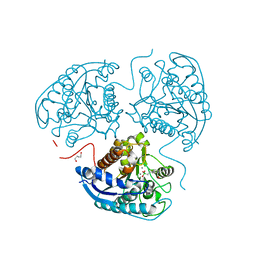 | | Structure of hARG1 with a novel inhibitor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[(1~{R},3~{R},4~{S})-3-azanyl-3-carboxy-4-[(dimethylamino)methyl]cyclohexyl]ethyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, ... | | Authors: | Napiorkowska-Gromadzka, A, Nowak, E, Nowotny, M. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Arginase 1/2 Inhibitor OATD-02: From Discovery to First-in-man Setup in Cancer Immunotherapy.
Mol.Cancer Ther., 22, 2023
|
|
7EU1
 
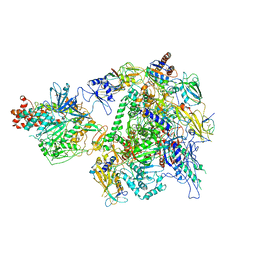 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 holoenzyme | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerases II and IV subunit 5A, DNA-directed RNA polymerases II, ... | | Authors: | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
7FB7
 
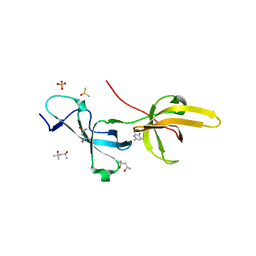 | | Crystal structure of human UHRF1 TTD in complex with 5-amino-2,4-dimethylpyridine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-amino-2,4-dimethylpyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Kori, S, Arita, K, Yoshimi, S. | | Deposit date: | 2021-07-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg.Med.Chem., 52, 2021
|
|
8G6D
 
 | |
8JRQ
 
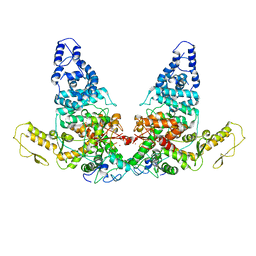 | | Structure of E6AP-E6 complex in Det1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
7F8L
 
 | |
6PJ2
 
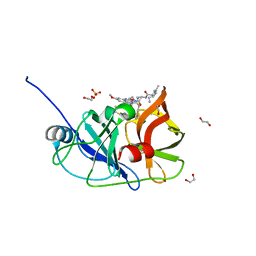 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-P5-4 (AJ-65) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-L-isoleucyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methylcyclo propyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacycl opentadecine-14a(5H)-carboxamide, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
8JRO
 
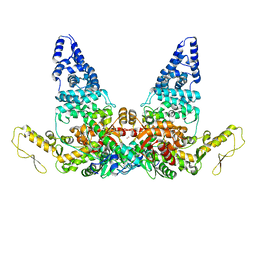 | | Structure of E6AP-E6 complex in Att2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
7EYK
 
 | |
7FC9
 
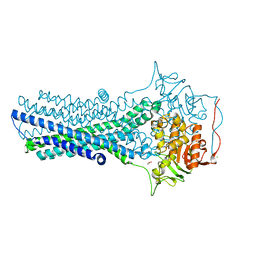 | | Crystal structure of CmABCB1 in lipidic mesophase revealed by LCP-SFX | | Descriptor: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pan, D, Oyama, R, Sato, T, Nakane, T, Mizunuma, R, Matsuoka, K, Joti, Y, Tono, K, Nango, E, Iwata, S, Nakatsu, T, Kato, H. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of CmABCB1 multi-drug exporter in lipidic mesophase revealed by LCP-SFX.
Iucrj, 9, 2022
|
|
7EYP
 
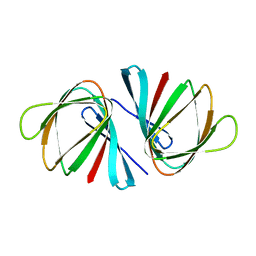 | | Crystal structure of Pseudomonas aeruginosa ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYJ
 
 | | Crystal structure of Escherichia coli ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase, SULFATE ION | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYL
 
 | | Crystal structure of Salmonella enterica ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYM
 
 | | Crystal structure of Vibrio cholerae ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
8JRN
 
 | | Structure of E6AP-E6 complex in Att1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
7FGV
 
 | |
7FG8
 
 | |
7FGU
 
 | |
7ERV
 
 | | Crystal structure of L-histidine decarboxylase (C57S/C101V/C282V mutant) from Photobacterium phosphoreum | | Descriptor: | Histidine decarboxylase, IMIDAZOLE | | Authors: | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
