4RVJ
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with amprenavir | | Descriptor: | HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2014-11-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1
To be Published
|
|
6NBD
 
 | |
6NBS
 
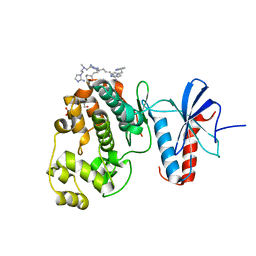 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
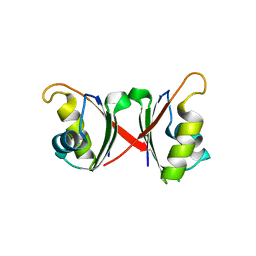
6NWW
 
 | |
4S34
 
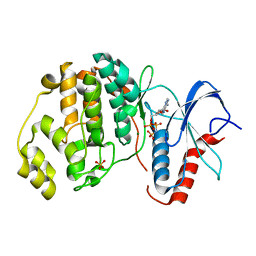 | | ERK2 (I84A) in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
4TMY
 
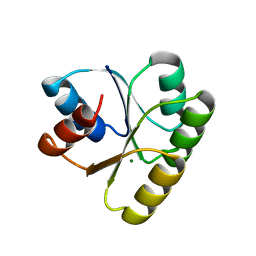 | | CHEY FROM THERMOTOGA MARITIMA (MG-IV) | | Descriptor: | CHEY PROTEIN, MAGNESIUM ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
4TRZ
 
 | | Structure of BACE1 complex with 2-thiophenyl HEA-type inhibitor | | Descriptor: | 2-thiophenyl HEA-type inhibitor, Beta-secretase 1 | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TSB
 
 | |
6NNX
 
 | |
4RTT
 
 | |
6NNY
 
 | |
4RYG
 
 | |
4RVT
 
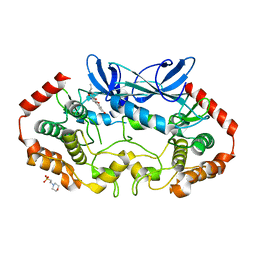 | | MAP4K4 in complex with a pyridin-2(1H)-one derivative | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-hexanoyl-4-hydroxy-5-(4-hydroxyphenyl)pyridin-2(1H)-one, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Richters, A, Becker, C, Kleine, S, Rauh, D. | | Deposit date: | 2014-11-27 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Neuritogenic Militarinone-Inspired 4-Hydroxypyridones Target the Stress Pathway Kinase MAP4K4.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6NFY
 
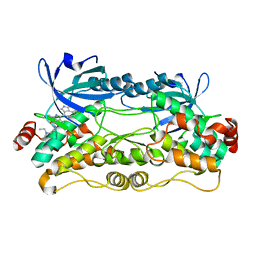 | | Crystal structure of nonphosphorylated, HPK1 kinase domain in complex with sunitinib in the inactive state. | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2018-12-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation.
J.Biol.Chem., 294, 2019
|
|
4S32
 
 | |
4S0Q
 
 | |
4SRN
 
 | | STRUCTURAL CHANGES THAT ACCOMPANY THE REDUCED CATALYTIC EFFICIENCY OF TWO SEMISYNTHETIC RIBONUCLEASE ANALOGS | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Martin, P.D, Doscher, M.S, Edwards, B.F.P. | | Deposit date: | 1991-05-20 | | Release date: | 1994-12-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs.
J.Biol.Chem., 267, 1992
|
|
6NWV
 
 | |
4TKB
 
 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with lauric acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TN1
 
 | |
4TRY
 
 | | Structure of BACE1 complex with a HEA-type inhibitor | | Descriptor: | Beta-secretase 1, GLU-ILE-TIH-THC-NVA | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of BACE1 complex with an anti-HMC-type inhibitor
to be published
|
|
4TU9
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5G6 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DG*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, McLaughlin, K.J, Agrawal, A.A, Kielkopf, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RKE
 
 | | Drosophila melanogaster Rab2 bound to GMPPNP | | Descriptor: | GH01619p, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Lardong, J.A, Driller, J.H, Depner, H, Weise, C, Petzoldt, A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | Structures of Drosophila melanogaster Rab2 and Rab3 bound to GMPPNP.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4RLD
 
 | |
4ROM
 
 | | Deoxyhemoglobin in complex with imidazolylacryloyl derivatives | | Descriptor: | 4-{2-chloro-4-[3-(1H-imidazol-2-yl)propanoyl]phenoxy}butanoic acid, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Omar, A.M, Mahran, M.A, Ghatge, M.N, Chowdhury, N, Bamane, F.H.A, El-Araby, M.E, Abdulmalik, O, Safo, M.K. | | Deposit date: | 2014-10-28 | | Release date: | 2015-05-20 | | Last modified: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a novel class of covalent modifiers of hemoglobin as potential antisickling agents.
Org.Biomol.Chem., 13, 2015
|
|
