6NJN
 
 | |
5HIS
 
 | |
5HK1
 
 | | Human sigma-1 receptor bound to PD144418 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-(4-methylphenyl)-5-(1-propyl-3,6-dihydro-2H-pyridin-5-yl)-1,2-oxazole, SULFATE ION, ... | | Authors: | Schmidt, H.R, Zheng, S, Gurpinar, E, Koehl, A, Manglik, A, Kruse, A.C. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5051 Å) | | Cite: | Crystal structure of the human sigma 1 receptor.
Nature, 532, 2016
|
|
5R0W
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 10, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1C
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 27, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1S
 
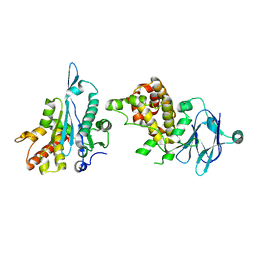 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 42, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7XZ0
 
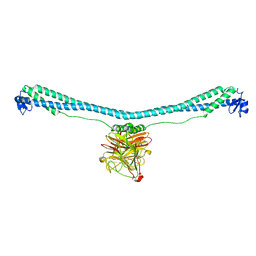 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5H5U
 
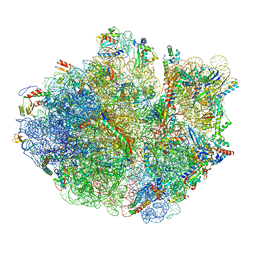 | | Mechanistic insights into the alternative translation termination by ArfA and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ma, C, Kurita, D, Li, N, Chen, Y, Himeno, H, Gao, N. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-25 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative translation termination by ArfA and RF2
Nature, 541, 2017
|
|
7ETL
 
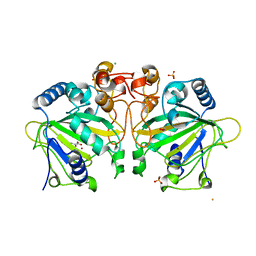 | | The crystal structure of FtmOx1-Y68F | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-05-13 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992128 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of the Endoperoxide Synthase FtmOx1.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y1H
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(5-chloranyl-4-methyl-benzimidazol-1-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
1HRB
 
 | |
3AIG
 
 | | ADAMALYSIN II WITH PEPTIDOMIMETIC INHIBITOR POL656 | | Descriptor: | (3R)-2-[N-(furan-2-ylcarbonyl)-L-leucyl]-2,3,4,9-tetrahydro-1H-beta-carboline-3-carboxylic acid, ADAMALYSIN II, CALCIUM ION, ... | | Authors: | Gomis-Rueth, F.X, Meyer, E.F, Kress, L.F, Politi, V. | | Deposit date: | 1997-10-12 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of adamalysin II with peptidic inhibitors. Implications for the design of tumor necrosis factor alpha convertase inhibitors.
Protein Sci., 7, 1998
|
|
5HA7
 
 | | Human Aldose Reductase in Complex with NADP+ and WY14643 in Space Group P212121 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sawaya, M.R, Cascio, D, Balendiran, G.K. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of WY 14,643 and its Complex with Aldose Reductase.
Sci Rep, 6, 2016
|
|
1THA
 
 | | MECHANISM OF MOLECULAR RECOGNITION. STRUCTURAL ASPECTS OF 3,3'-DIIODO-L-THYRONINE BINDING TO HUMAN SERUM TRANSTHYRETIN | | Descriptor: | 3,3'-DEIODO-THYROXINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Luft, J, Cody, V. | | Deposit date: | 1991-11-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of molecular recognition. Structural aspects of 3,3'-diiodo-L-thyronine binding to human serum transthyretin.
J.Biol.Chem., 267, 1992
|
|
6MR8
 
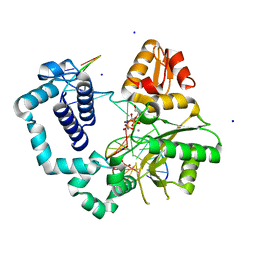 | | D276G DNA polymerase beta substrate complex with templating adenine and incoming Fapy-dGTP analog | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
7XQ5
 
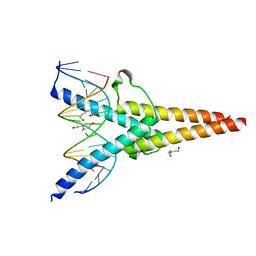 | | Crystal structure of ScIno2p-ScIno4p bound promoter DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*TP*CP*AP*CP*AP*TP*GP*CP*AP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*AP*TP*GP*TP*GP*AP*AP*AP*AP*T)-3'), HEXANE-1,6-DIOL, ... | | Authors: | Khan, M.H, Lu, X. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA.
Int J Mol Sci, 23, 2022
|
|
4ABF
 
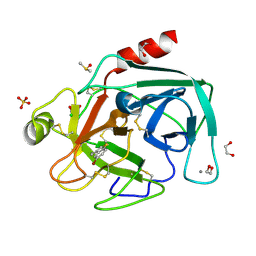 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-BROMO-1-BENZOTHIOPHEN-3-YL)METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
5H1T
 
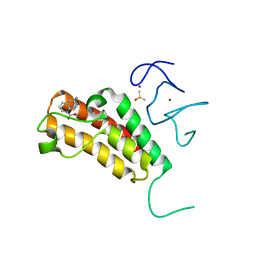 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 1 | | Descriptor: | DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ZINC ION, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
6GVQ
 
 | |
7Y8V
 
 | | Crystal structure of AlbEF homolog mutant (AlbF-H54A/H58A) from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog H54A/H58A mutant, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8U
 
 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8X
 
 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans in complex with Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
1VCC
 
 | |
7XQN
 
 | |
7XQM
 
 | |
