5BXN
 
 | | Yeast 20S proteasome beta2-G170A mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
5BYG
 
 | | X-ray structure of AAV2 OBD-AAVS1 complex 2:1 | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Musayev, F.N, Escalante, C.R. | | Deposit date: | 2015-06-10 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Assembly of the Adeno-associated Virus Type 2 Rep68 Protein on the Integration Site AAVS1.
J.Biol.Chem., 290, 2015
|
|
2DPD
 
 | |
2MW7
 
 | |
2NQO
 
 | | Crystal Structure of Helicobacter pylori gamma-Glutamyltranspeptidase | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Boanca, G, Sand, A, Okada, T, Suzuki, H, Kumagai, H, Fukuyama, K, Barycki, J.J. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad.
J.Biol.Chem., 282, 2007
|
|
6ALH
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex | | Descriptor: | DNA (29-MER), DNA (5'-D(*GP*GP*GP*CP*TP*AP*AP*TP*GP*AP*CP*GP*GP*CP*GP*AP*AP*TP*AP*CP*CP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Darst, S.A. | | Deposit date: | 2017-08-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of transcription arrest by coliphage HK022 Nun in anEscherichia coliRNA polymerase elongation complex.
Elife, 6, 2017
|
|
5CGI
 
 | | Yeast 20S proteasome beta5-G48C mutant in complex with ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dubiella, C, Groll, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective Inhibition of the Immunoproteasome by Structure-Based Targeting of a Non-catalytic Cysteine.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5CIO
 
 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
6ATU
 
 | |
6APD
 
 | | Crystal structure of RSV F bound by AM22 and the infant antibody ADI-19425 | | Descriptor: | AM22 Fab Heavy Chain,IGH@ protein, AM22 Fab Light Chain,Uncharacterized protein, Fusion glycoprotein F0,Envelope glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation.
Immunity, 48, 2018
|
|
5C0T
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
6AUI
 
 | | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Brignole, E.J, Drennan, C.L, Asturias, F.J, Tsai, K.L, Penczek, P.A. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3- angstrom resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound.
Elife, 7, 2018
|
|
5C2N
 
 | | The de novo evolutionary emergence of a symmetrical protein is shaped by folding constraints | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta propeller | | Authors: | Smock, R.G, Yadid, I, Dym, O, Clarke, J, Tawfik, D.S. | | Deposit date: | 2015-06-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De Novo Evolutionary Emergence of a Symmetrical Protein Is Shaped by Folding Constraints.
Cell, 164, 2016
|
|
6ATF
 
 | | HLA-DRB1*1402 in complex with Vimentin59-71 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scally, S.W, Ting, Y.T, Rossjohn, J. | | Deposit date: | 2017-08-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis.
Ann. Rheum. Dis., 76, 2017
|
|
2NNW
 
 | | Alternative conformations of Nop56/58-fibrillarin complex and implication for induced-fit assenly of box C/D RNPs | | Descriptor: | Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein | | Authors: | Oruganti, S, Zhang, Y, Terns, R, Terns, M.P, Li, H. | | Deposit date: | 2006-10-24 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Conformations of the Archaeal Nop56/58-Fibrillarin Complex Imply Flexibility in Box C/D RNPs.
J.Mol.Biol., 371, 2007
|
|
6B4P
 
 | |
5CGF
 
 | | Yeast 20S proteasome beta5-G48C mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Dubiella, C, Groll, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective Inhibition of the Immunoproteasome by Structure-Based Targeting of a Non-catalytic Cysteine.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2EFW
 
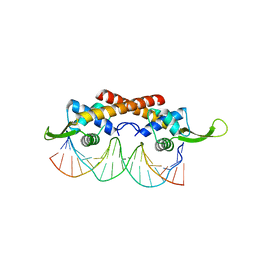 | | Crystal structure of the RTP:nRB complex from Bacillus subtilis | | Descriptor: | DNA (5'-D(*DCP*DT*DAP*DTP*DGP*DTP*DAP*DCP*DCP*DAP*DAP*DAP*DTP*DGP*DTP*DTP*DCP*DAP*DGP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DCP*DTP*DGP*DAP*DAP*DCP*DAP*DTP*DTP*DTP*DGP*DGP*DTP*DAP*DCP*DAP*DTP*DAP*DG)-3'), Replication termination protein | | Authors: | Vivian, J.P, Porter, C.J, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2007-02-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA
J.Mol.Biol., 370, 2007
|
|
6B83
 
 | |
2NNB
 
 | | The Q403K mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Anderson, J.L.R, Miles, C.S, Mowat, C.G, Reid, G.A, Chapman, S.K. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Q403K mutant heme domain of flavocytochrome P450 BM3
To be published
|
|
6BAA
 
 | | Cryo-EM structure of the pancreatic beta-cell KATP channel bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Martin, G.M, Yoshioka, C, Shyng, S.L. | | Deposit date: | 2017-10-12 | | Release date: | 2017-11-01 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Elife, 6, 2017
|
|
5C0W
 
 | |
2NN6
 
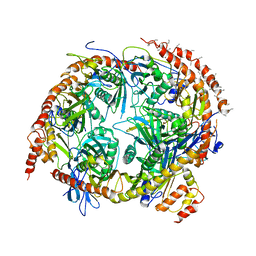 | | Structure of the human RNA exosome composed of Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, Rrp42, Csl4, Rrp4, and Rrp40 | | Descriptor: | 3'-5' exoribonuclease CSL4 homolog, Exosome complex exonuclease RRP4, Exosome complex exonuclease RRP40, ... | | Authors: | Lima, C.D. | | Deposit date: | 2006-10-23 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Reconstitution, activities, and structure of the eukaryotic RNA exosome.
Cell(Cambridge,Mass.), 127, 2006
|
|
6ATZ
 
 | | HLA-DRB1*1402 in complex with citrullinated fibrinogen peptide | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta chain, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-08-29 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis.
Ann. Rheum. Dis., 76, 2017
|
|
5C80
 
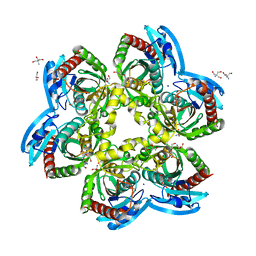 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
