7XN5
 
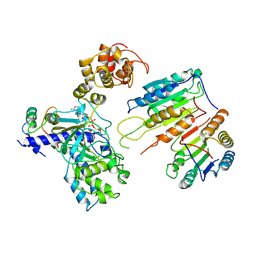 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
8TRG
 
 | |
5NQ7
 
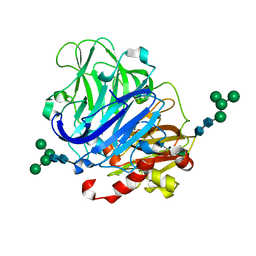 | | Crystal structure of laccases from Pycnoporus sanguineus, izoform I | | Descriptor: | COPPER (II) ION, Laccase, PEROXIDE ION, ... | | Authors: | Orlikowska, M, de J.Rostro-Alanis, M, Bujacz, A, Hernandez-Luna, C, Rubio, R, Parra, R, Bujacz, G. | | Deposit date: | 2017-04-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies of two thermostable laccases from the white-rot fungus Pycnoporus sanguineus.
Int. J. Biol. Macromol., 107, 2018
|
|
1LI1
 
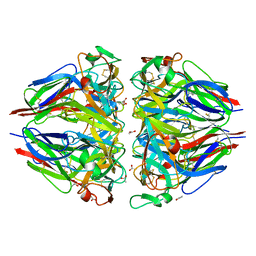 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2KS9
 
 | |
6B8L
 
 | |
6XN6
 
 | |
7MJ0
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with adenosine monophosphate AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2KQX
 
 | |
7MJ1
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with NAD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJ2
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zn | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5NQ9
 
 | | Crystal structure of laccases from Pycnoporus sanguineus, izoform II, monoclinic | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Orlikowska, M, de J.Rostro-Alanis, M, Bujacz, A, Hernandez-Luna, C, Rubio, R, Parra, R, Bujacz, G. | | Deposit date: | 2017-04-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural studies of two thermostable laccases from the white-rot fungus Pycnoporus sanguineus.
Int. J. Biol. Macromol., 107, 2018
|
|
1JFM
 
 | |
2KSB
 
 | |
5NOC
 
 | |
5L7J
 
 | |
6XPA
 
 | |
3R66
 
 | | Crystal structure of human ISG15 in complex with NS1 N-terminal region from influenza virus B, Northeast Structural Genomics Consortium Target IDs HX6481, HR2873, and OR2 | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Guan, R, Ma, L.C, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the complex of ISG15 and Influenza B virus NS1 Protein: implications for the mechanism of SN1-mediated inhibition of ISG15 conjugation
To be Published
|
|
2GPE
 
 | |
7BJP
 
 | | The cryo-EM structure of vesivirus 2117, an adventitious agent and possible cause of haemorrhagic gastroenteritis in dogs. | | Descriptor: | Capsid protein | | Authors: | Sutherland, H, Conley, M.J, Emmott, E, Streetley, J, Goodfellow, I.G, Bhella, D. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The Cryo-EM Structure of Vesivirus 2117 Highlights Functional Variations in Entry Pathways for Viruses in Different Clades of the Vesivirus Genus.
J.Virol., 95, 2021
|
|
5L7L
 
 | |
6DAF
 
 | | 2.4 Angstrom crystal structure of the F141L Ca/CaM:CaV1.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1JUH
 
 | | Crystal Structure of Quercetin 2,3-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fusetti, F, Schroeter, K.H, Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-08-24 | | Release date: | 2002-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus.
Structure, 10, 2002
|
|
6C74
 
 | |
7BFL
 
 | | X-ray structure of SS-RNase-2 des116-120 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
