2JQ2
 
 | |
5JBN
 
 | |
1ZRY
 
 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
8DGH
 
 | |
8DGK
 
 | |
8DFZ
 
 | |
1Q9F
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX.
J.Mol.Biol., 336, 2004
|
|
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
8VSX
 
 | | NMR Structure of GCAP5 R22A | | Descriptor: | Guanylyl cyclase-activating protein 1 | | Authors: | Cudia, D.L, Ames, J.B. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Retinal Guanylate Cyclase Activating Protein 5 (GCAP5) with R22A Mutation That Abolishes Dimerization and Enhances Cyclase Activation.
Biochemistry, 63, 2024
|
|
6OSO
 
 | | The crystal structure of the isolate tryptophan synthase alpha-chain from Salmonella enterica serovar typhimurium at 1.75 Angstrom resolution | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L, Chang, C, Fan, L. | | Deposit date: | 2019-05-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Backbone assignments and conformational dynamics in the S. typhimurium tryptophan synthase alpha-subunit from solution-state NMR.
J.Biomol.Nmr, 74, 2020
|
|
6OUY
 
 | | The crystal structure of the isolate tryptophan synthase alpha-chain from Salmonella enterica serovar typhimurium at 1.60 Angstrom resolution | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L, Chang, C, Fan, L. | | Deposit date: | 2019-05-06 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Backbone assignments and conformational dynamics in the S. typhimurium tryptophan synthase alpha-subunit from solution-state NMR.
J.Biomol.Nmr, 74, 2020
|
|
7DFE
 
 | | NMR structure of TuSp2-RP | | Descriptor: | B6 protein | | Authors: | Lin, Z, Fan, T, Fan, J. | | Deposit date: | 2020-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments of a repetitive domain of tubuliform spidroin 2
Biomol.Nmr Assign., 15, 2021
|
|
7MQU
 
 | | The haddock model of GDP KRas in complex with promethazine using NMR chemical shift perturbations | | Descriptor: | (2R)-N,N-dimethyl-1-(10H-phenothiazin-10-yl)propan-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.P. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
7AQT
 
 | | NMR2 structure of BRD4-BD2 in complex with iBET-762 | | Descriptor: | 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide, Bromodomain-containing protein 4 | | Authors: | Orts, J, Torres, F, Milbradt, A.G, Walser, R. | | Deposit date: | 2020-10-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Molecular Replacement Provides New Insights into Binding Modes to Bromodomains of BRD4 and TRIM24.
J.Med.Chem., 65, 2022
|
|
7YVW
 
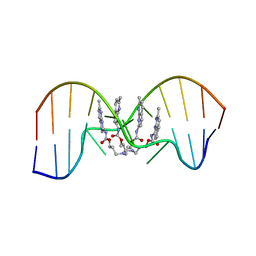 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
7M2M
 
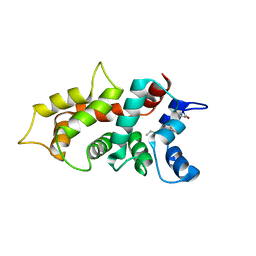 | | NMR Structure of GCAP5 | | Descriptor: | Guanylate cyclase activator 1A, MAGNESIUM ION | | Authors: | Ames, J.B, Cudia, D.L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR and EPR-DEER Structure of a Dimeric Guanylate Cyclase Activator Protein-5 from Zebrafish Photoreceptors.
Biochemistry, 60, 2021
|
|
7B9X
 
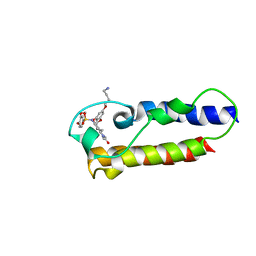 | | NMR2 structure of TRIM24-BD in complex with a precursor of IACS-9571 | | Descriptor: | N-{6-[3-(4-Aminobutoxy)-5-propoxyphenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide, Transcription intermediary factor 1-alpha | | Authors: | Orts, J, Torres, F, Milbradt, A.G, Walser, R. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Molecular Replacement Provides New Insights into Binding Modes to Bromodomains of BRD4 and TRIM24.
J.Med.Chem., 65, 2022
|
|
6LUR
 
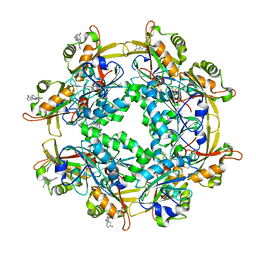 | |
6SOW
 
 | |
6T51
 
 | |
8SXM
 
 | |
5ZFO
 
 | | NMR structure of IRD12 from Capsicum annum. | | Descriptor: | Pin-II type proteinase inhibitor 38 | | Authors: | Gartia, J, Barnwal, R.P, Chary, K.V.R. | | Deposit date: | 2018-03-06 | | Release date: | 2019-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of inhibitory repeat domain variant 12, a plant protease inhibitor from Capsicum annuum, and its structural relationship to other plant protease inhibitors.
J.Biomol.Struct.Dyn., 2019
|
|
7KW9
 
 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
7KW8
 
 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis | | Descriptor: | tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
6V1W
 
 | |
