6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagY | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6S6V
 
 | | Resting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ATPgS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Nuclease SbcCD subunit C, ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-04 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
6L0L
 
 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
6RHL
 
 | | Room temperature data of Galectin-3C in complex with a pair of enantiomeric ligands: R enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{R})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Are crystallographic B-factors suitable for calculating protein conformational entropy?
Phys Chem Chem Phys, 21, 2019
|
|
7AC1
 
 | | Solution structure of the TAF4-RST domain | | Descriptor: | Transcription initiation factor TFIID subunit 4 | | Authors: | Staby, L, Davidsen, R, Bugge, K, Skriver, K, Kragelund, B.B. | | Deposit date: | 2020-09-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | alpha alpha-hub coregulator structure and flexibility determine transcription factor binding and selection in regulatory interactomes.
J.Biol.Chem., 298, 2022
|
|
7PZJ
 
 | | Structure of a bacteroidetal polyethylene terephthalate (PET) esterase | | Descriptor: | Lipase, POTASSIUM ION | | Authors: | Zang, H, Dierkes, R, Perez-Garcia, P, Weigert, S, Sternagel, S, Hallam, S.J, Applegate, V, Schumacher, J, Schott, T, Pleiss, J, Almeida, A, Hoecker, B, Smits, S.H, Schmitz, R.A, Chow, J, Streit, W.R. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity.
Front Microbiol, 12, 2021
|
|
7Q4Y
 
 | |
7N34
 
 | |
6RHM
 
 | | Room temperature data of Galectin-3C in complex with a pair of enantiomeric ligands: S enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{S})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Are crystallographic B-factors suitable for calculating protein conformational entropy?
Phys Chem Chem Phys, 21, 2019
|
|
5DZ1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-[(4-ethylcyclohexylidene)methanediyl]diphenol | | Descriptor: | 4,4'-[(4-ethylcyclohexylidene)methanediyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7JTZ
 
 | | Yeast Glo3 GAP domain | | Descriptor: | ADP-ribosylation factor GTPase-activating protein GLO3, GLYCEROL, ZINC ION | | Authors: | Xie, B, Jackson, L.P. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Glo3 GAP crystal structure supports the molecular niche model for ArfGAPs in COPI coats.
Adv Biol Regul, 79, 2021
|
|
5ZUR
 
 | | Achromobacter Dh1f Bacterioferritin | | Descriptor: | BARIUM ION, Bacterioferritin, CHLORIDE ION, ... | | Authors: | Dwivedy, A, Jha, B, Singh, K.H, Ahmed, M, Ashraf, A, Kumar, D, Biswal, B.K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous crystallization and structure determination of bacterioferritin from Achromobacter.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7Q2X
 
 | | Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (Form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Condensin complex subunit 1, ... | | Authors: | Lee, B.-G, Rhodes, J, Lowe, J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Clamping of DNA shuts the condensin neck gate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NAU
 
 | | Bacterial 30S ribosomal subunit assembly complex state C (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAV
 
 | | Bacterial 30S ribosomal subunit assembly complex state D (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7PLM
 
 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
3NKB
 
 | | A 1.9A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage | | Descriptor: | DNA/RNA (5'-D(*(DUR))-D(*GP*G)-R(P*CP*UP*UP*GP*CP*A)-3'), MAGNESIUM ION, The hepatitis delta virus ribozyme | | Authors: | Chen, J.-H, Yajima, R, Chadalavada, D.M, Chase, E, Bevilacqua, P.C, Golden, B.L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage.
Biochemistry, 49, 2010
|
|
7Q2Y
 
 | | Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (form II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Condensin complex subunit 1, ... | | Authors: | Lee, B.-G, Rhodes, J, Lowe, J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-23 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Clamping of DNA shuts the condensin neck gate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NAT
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAX
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state I (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAR
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state F (+RsgA)(Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
6RP2
 
 | | Threonine to Cysteine (T225C) variant of E coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L. | | Deposit date: | 2019-05-13 | | Release date: | 2020-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Aerobic Photocatalytic H2Production by a [NiFe] Hydrogenase Engineered to Place a Silver Nanocluster in the Electron Relay.
J.Am.Chem.Soc., 142, 2020
|
|
7NK0
 
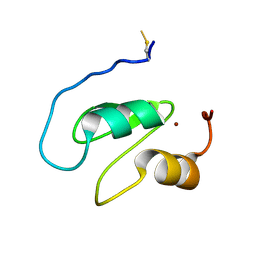 | | Structure of the BIR1 domain of cIAP2 | | Descriptor: | Baculoviral IAP repeat-containing protein 3, ZINC ION | | Authors: | Cossu, F, Milani, M, Mastrangelo, E, Mirdita, D. | | Deposit date: | 2021-02-17 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based identification of a new IAP-targeting compound that induces cancer cell death inducing NF-kappa B pathway.
Comput Struct Biotechnol J, 19, 2021
|
|
7Q2Z
 
 | |
4PET
 
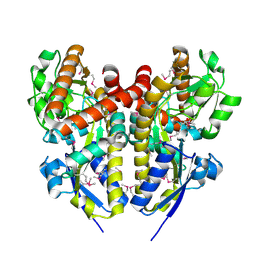 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM COLWELLIA PSYCHRERYTHRAEA (CPS_0129, TARGET EFI-510097) WITH BOUND CALCIUM AND PYRUVATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
