8U44
 
 | | CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex with 05.GC.w2.3C10-H1_SI06 | | Descriptor: | 05.GC.w2.3C10-H1_SI06 Heavy chain, 05.GC.w2.3C10-H1_SI06 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moore, N, Han, J, Ward, A.B, Wilson, I.A. | | Deposit date: | 2023-09-08 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Persistence of germinal center B cell responses after influenza virus vaccination in humans
To Be Published
|
|
8BIP
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
7H2W
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z3765495818 (A71EV2A-x0202) | | Descriptor: | (4R)-imidazo[1,2-b]pyridazine-3-sulfonamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7A5P
 
 | | Human C Complex Spliceosome - Medium-resolution PERIPHERY | | Descriptor: | Cell division cycle 5-like protein, Crooked neck-like protein 1, Eukaryotic initiation factor 4A-III, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
7GZI
 
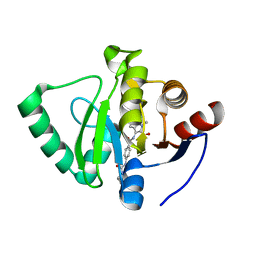 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000753-001 | | Descriptor: | 3-[4-(cyclopropylcarbamamido)benzamido]-1-methyl-1H-pyrrolo[2,3-b]pyridine-2-carboxylic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZF
 
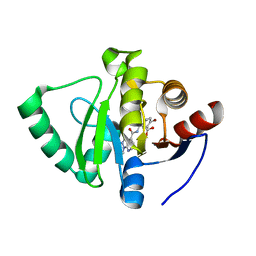 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000605-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1-methyl-1H-pyrazolo[3,4-b]pyridine-4-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZH
 
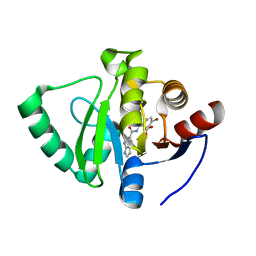 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000670-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1H-pyrrolo[2,3-b]pyridine-5-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZG
 
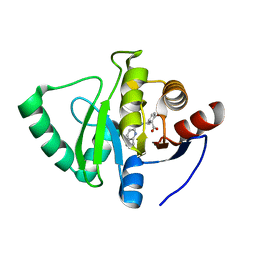 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000620-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1H-pyrrolo[3,2-b]pyridine-5-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6WY5
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-37 A.K.A 7-(1-phenyl-3-(((1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl)amino)propyl)-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1-phenyl-3-{[(1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6QZP
 
 | | High-resolution cryo-EM structure of the human 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S rRNA (1740-MER), 28S rRNA (3773-MER), ... | | Authors: | Natchiar, S.K, Myasnikov, A.G, Kratzat, H, Hazemann, I, Klaholz, B.P. | | Deposit date: | 2019-03-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualization of chemical modifications in the human 80S ribosome structure.
Nature, 551, 2017
|
|
7H0B
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0010716-001 | | Descriptor: | (4M)-4-(4-{[(1R)-1-(2,3-dihydro[1,4]dioxino[2,3-b]pyridin-6-yl)-2-methylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1-methyl-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6H9F
 
 | | Structure of glutamate mutase reconstituted with bishomo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5M39
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 1 | | Descriptor: | 6-(3,4-dimethoxyphenyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-12-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
6N1L
 
 | | The complement inhibitory domain of B. burgdorferi BBK32. | | Descriptor: | Fibronectin-binding protein BBK32 | | Authors: | Garcia, B.L. | | Deposit date: | 2018-11-09 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Structural determination of the complement inhibitory domain of Borrelia burgdorferi BBK32 provides insight into classical pathway complement evasion by lyme disease spirochetes.
PLoS Pathog., 15, 2019
|
|
5V54
 
 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
8KC9
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to cyclosporin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, DSN-MLE-MLE-MVA-BMT-ABA-SAR-MLE-VAL-MLE-ALA, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0M
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-09 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0F
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K17
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to collagen alpha-1(I) chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-10 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8V4X
 
 | | Structure of MALT1 in complex with an allosteric inhibitor | | Descriptor: | Inhibitor peptide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, N-{7-[(1S)-1-methoxyethyl]-2-methyl[1,3]thiazolo[5,4-b]pyridin-6-yl}-N'-[6-(2H-1,2,3-triazol-2-yl)-5-(trifluoromethyl)pyridin-3-yl]urea | | Authors: | Judge, R.A, Pappano, W.N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Inhibition of MALT1 and BCL2 Induces Synergistic Antitumor Activity in Models of B-Cell Lymphoma.
Mol.Cancer Ther., 23, 2024
|
|
4RVM
 
 | | CHK1 kinase domain with diazacarbazole compound 19 | | Descriptor: | 3-[4-(piperidin-1-ylmethyl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Gazzard, L, Blackwood, E, Burton, B, Chapman, K, Chen, H, Crackett, P, Drobnick, J, Ellwood, C, Epler, J, Flagella, M, Goodacre, S, Halladay, J, Hunt, H, Kintz, S, Lyssikatos, J, MacLeod, C, Ramiscal, S, Schmidt, S, Seward, E, Wiesmann, C, Williams, K, Wu, P, Yee, S, Yen, I, Malek, S. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
6CVK
 
 | |
7JYU
 
 | | Crystal Structure of HLA-A*2402 in complex with IYFSPIRVTF, an 10-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Nguyen, A.T, Szeto, C, Rossjohn, J, Gras, S. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7Z43
 
 | |
