5EL2
 
 | |
5DNC
 
 | |
8U8J
 
 | |
8U8K
 
 | |
8TTN
 
 | | PHF1-Phosphomimetic Tau Filaments (Full-length, Cofactor-Free 0N4R Tau S396E, S400E, T403E, S404E) | | Descriptor: | Microtubule-associated protein tau | | Authors: | El Mammeri, N, Dregni, A.J, Duan, P, Hong, M. | | Deposit date: | 2023-08-14 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of AT8 and PHF1 phosphomimetic tau: Insights into the posttranslational modification code of tau aggregation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TTL
 
 | | AT8-Phosphomimetic Tau Filaments (Full-length, Cofactor-Free 0N4R Tau S202E, T205E, S208E) | | Descriptor: | Microtubule-associated protein tau | | Authors: | El Mammeri, N, Dregni, A.J, Duan, P, Hong, M. | | Deposit date: | 2023-08-14 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of AT8 and PHF1 phosphomimetic tau: Insights into the posttranslational modification code of tau aggregation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4TW3
 
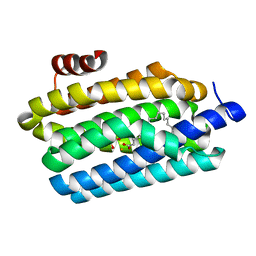 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
3DGD
 
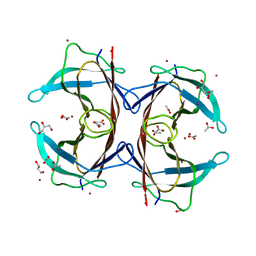 | | Crystal structure of the F87M/L110M mutant of human transthyretin at pH 4.6 | | Descriptor: | ACETATE ION, GLYCEROL, Transthyretin, ... | | Authors: | Palmieri, L.C, Freire, J.B.B, Foguel, D, Lima, L.M.T.R. | | Deposit date: | 2008-06-13 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Novel Zn2+-binding sites in human transthyretin: implications for amyloidogenesis and retinol-binding protein recognition.
J.Biol.Chem., 285, 2010
|
|
4UF8
 
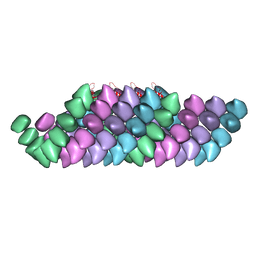 | | Electron cryo-microscopy structure of PB1-p62 filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
4PKY
 
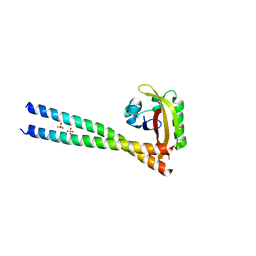 | | ARNT/HIF transcription factor/coactivator complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, SULFATE ION, ... | | Authors: | Tomchick, D.R, Partch, C.L, Gardner, K.H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coiled-coil Coactivators Play a Structural Role Mediating Interactions in Hypoxia-inducible Factor Heterodimerization.
J.Biol.Chem., 290, 2015
|
|
4PX9
 
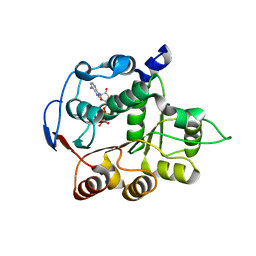 | | DEAD-box RNA helicase DDX3X Domain 1 with N-terminal ATP-binding Loop | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX3X | | Authors: | Epling, L.B, Grace, C.R, Lowe, B.R, Partridge, J.F, Enemark, E.J. | | Deposit date: | 2014-03-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cancer-Associated Mutants of RNA Helicase DDX3X Are Defective in RNA-Stimulated ATP Hydrolysis.
J.Mol.Biol., 427, 2015
|
|
4QFQ
 
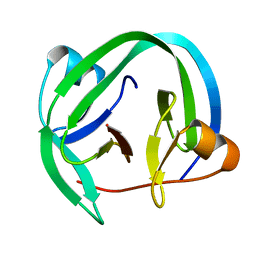 | |
4PHX
 
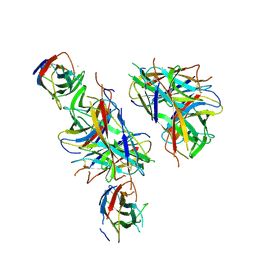 | |
4PH8
 
 | |
2Y6X
 
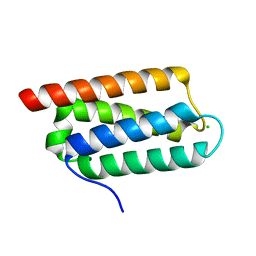 | | Structure of Psb27 from Thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, PHOTOSYSTEM II 11 KD PROTEIN | | Authors: | Michoux, F, Takasaka, K, Boehm, M, Nixon, P.J, Murray, J.W. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Psb27 Assembly Factor at 1.6A: Implications for Binding to Photosystem II.
Photosynth.Res., 110, 2012
|
|
4R71
 
 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | Descriptor: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | Deposit date: | 2014-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
5CTX
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPH
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTW
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(butanoylamino)thiophene-3-carboxamide, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Hadfield, A.T, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPV
 
 | |
5DDP
 
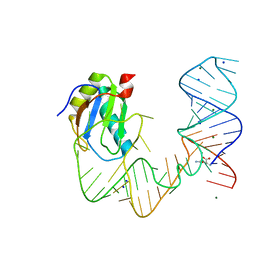 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
4R0E
 
 | |
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CRW
 
 | |
8KG3
 
 | | Structure of THOUSAND-GRAIN WEIGHT 6 (TGW6) | | Descriptor: | Os06g0623700 protein | | Authors: | Akabane, T, Suzuki, N, Matsumura, H, Yoshizawa, T, Tsuchiya, W, Katoh, E, Hirotsu, N. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | THOUSAND-GRAIN WEIGHT 6, which is an IAA-glucose hydrolase, preferentially recognizes the structure of the indole ring.
Sci Rep, 14, 2024
|
|
