2JF6
 
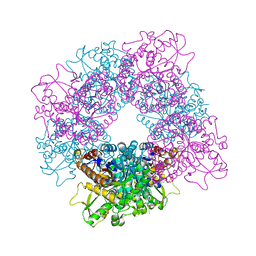 | | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
1QL8
 
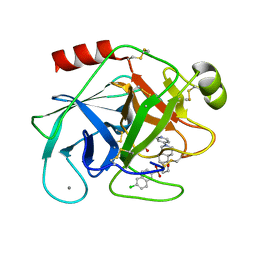 | |
2JED
 
 | | The crystal structure of the kinase domain of the protein kinase C theta in complex with NVP-XAA228 at 2.32A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(8-DIMETHYLAMINOMETHYL-6,7,8,9-TETRAHYDRO-PYRIDO[1,2-A]INDOL-10-YL)-4-(1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, PROTEIN KINASE C THETA | | Authors: | Stark, W, Bitsch, F, Berner, A, Buelens, F, Graff, P, Depersin, H, Geiser, M, Knecht, R, Rahuel, J, Rummel, G, Schlaeppi, J.M, Schmitz, R, Strauss, A, Wagner, J. | | Deposit date: | 2007-01-16 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Crystal Structure of the Kinase Domain of the Protein Kinase C Theta in Complex with Nvp-Xaa228
To be Published
|
|
1YW8
 
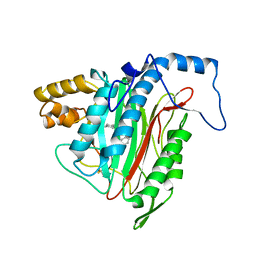 | | h-MetAP2 complexed with A751277 | | Descriptor: | 2-[(PHENYLSULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
1MGY
 
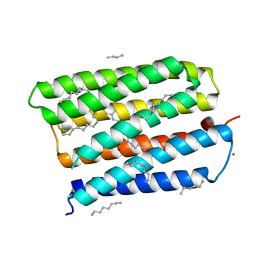 | | Structure of the D85S mutant of bacteriorhodopsin with bromide bound | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, BROMIDE ION, Bacteriorhodopsin, ... | | Authors: | Facciotti, M.T, Cheung, V.S, Nguyen, D, Rouhani, S, Glaeser, R.M. | | Deposit date: | 2002-08-16 | | Release date: | 2003-07-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bromide-Bound D85S Mutant of Bacteriorhodopsin:
Principles of Ion Pumping
Biophys.J., 85, 2003
|
|
1M00
 
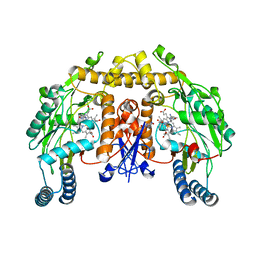 | | Rat neuronal NOS heme domain with N-butyl-N'-hydroxyguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-BUTYL-N'-HYDROXYGUANIDINE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
1YZ3
 
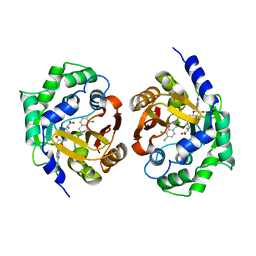 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
1YW9
 
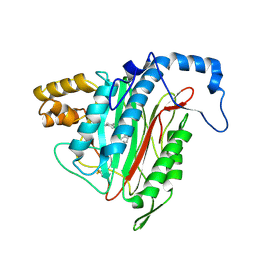 | | h-MetAP2 complexed with A849519 | | Descriptor: | 2-[({2-[(1Z)-3-(DIMETHYLAMINO)PROP-1-ENYL]-4-FLUOROPHENYL}SULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
2D2C
 
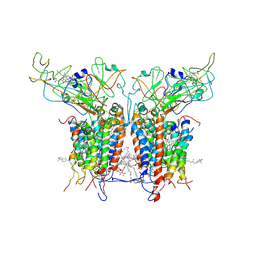 | | Crystal Structure Of Cytochrome B6F Complex with DBMIB From M. Laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, Apocytochrome f, ... | | Authors: | Yan, J, Kurisu, G, Cramer, W.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-12-13 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2K4L
 
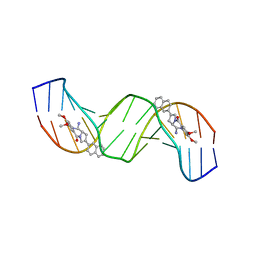 | | Solution structure of a 2:1C2-(2-naphthyl)pyrrolo[2,1-c][1,4]benzodiazepine (PBD) DNA adduct: molecular basis for unexpectedly high DNA helix stabilization. | | Descriptor: | (11aS)-7,8-dimethoxy-2-naphthalen-2-yl-1,10,11,11a-tetrahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DAP*DAP*DAP*DGP*DAP*DTP*DT)-3' | | Authors: | Antonow, D, Barata, T, Jenkins, T.C, Parkinson, G.N, Howard, P.W, Thurston, D.E, Zloh, M. | | Deposit date: | 2008-06-13 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a 2:1 C2-(2-naphthyl) pyrrolo[2,1-c][1,4]benzodiazepine DNA adduct: molecular basis for unexpectedly high DNA helix stabilization.
Biochemistry, 47, 2008
|
|
1YV3
 
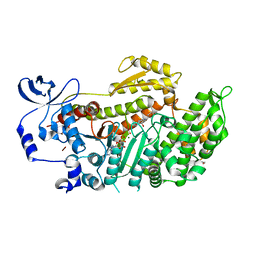 | | The structural basis of blebbistatin inhibition and specificity for myosin II | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Smith, R, Rayment, I. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of blebbistatin inhibition and specificity for myosin II.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2K8U
 
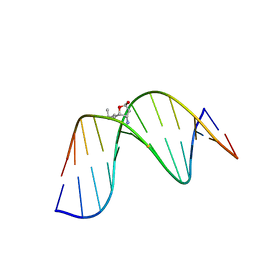 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6S,8R,11S)-configuration matched with dC | | Descriptor: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
2FUD
 
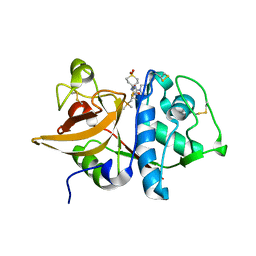 | | Human Cathepsin S with Inhibitor CRA-27566 | | Descriptor: | N-{(1R)-2-[(4-CYANO-1,1-DIOXIDOTETRAHYDRO-2H-THIOPYRAN-4-YL)AMINO]-2-OXO-1-[(TRIMETHYLSILYL)METHYL]ETHYL}MORPHOLINE-4-CARBOXAMIDE, cathepsin S | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Cathepsin S with Inhibitor CRA-27566
To be Published
|
|
2AB8
 
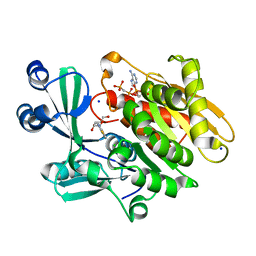 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside and AMP-PCP | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2FTD
 
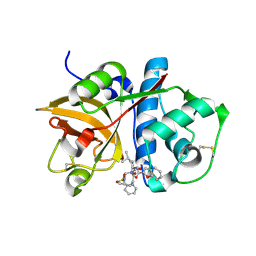 | | Crystal structure of Cathepsin K complexed with 7-Methyl-Substituted Azepan-3-one compound | | Descriptor: | Cathepsin K, N-[(1S)-1-({[(3S,4S,7R)-3-HYDROXY-7-METHYL-1-(PYRIDIN-2-YLSULFONYL)-2,3,4,7-TETRAHYDRO-1H-AZEPIN-4-YL]AMINO}CARBONYL)-3-METHYLBUTYL]-1-BENZOFURAN-2-CARBOXAMIDE | | Authors: | Yamashita, D.S, Baoguang, Z. | | Deposit date: | 2006-01-24 | | Release date: | 2007-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure activity relationships of 5-, 6-, and 7-methyl-substituted azepan-3-one cathepsin K inhibitors.
J.Med.Chem., 49, 2006
|
|
2FQI
 
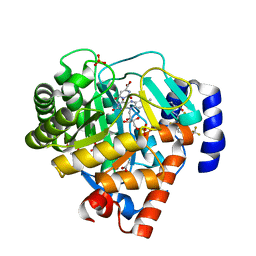 | | dual binding modes of a novel series of DHODH inhibitors | | Descriptor: | 2-({[2,3,5,6-TETRAFLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENTA-1,3-DIENE-1-CARBOXYLIC ACID, ACETATE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Baumgartner, R, Leban, J. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J.Med.Chem., 49, 2006
|
|
2FRQ
 
 | | Human Cathepsin S with Inhibitor CRA-26871 | | Descriptor: | N-[4-(AMINOMETHYL)-1,1-DIOXIDOTETRAHYDRO-2H-THIOPYRAN-4-YL]-3-(1-METHYLCYCLOPENTYL)-N~2~-[(1E)-N-(PHENYLSULFONYL)ETHANIMIDOYL]-L-ALANINAMIDE, cathepsin S | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-19 | | Release date: | 2006-07-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Cathepsin S with Inhibitor CRA-26871
To be Published
|
|
1E5J
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH METHYL-4II-S-ALPHA-CELLOBIOSYL-4II-THIO-BETA-CELLOBIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Fort, S, Varrot, A, Schulein, M, Cottaz, S, Driguez, H, Davies, G.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mixed-Linkage Cellooligosaccharides: A New Class of Glycoside Hydrolase Inhibitors
Chembiochem, 2, 2001
|
|
2A9B
 
 | | Crystal structure of R138Q mutant of recombinant sulfite oxidase at resting state | | Descriptor: | CHLORIDE ION, MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2AI2
 
 | | Purine nucleoside phosphorylase from calf spleen | | Descriptor: | ((2S,3AS,4R,6S)-4-(HYDROXYMETHYL)-6-(4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-TETRAHYDROFURO[3,4-D][1,3]DIOXO L-2-YL)METHYLPHOSPHONIC ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | Deposit date: | 2005-07-28 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
143D
 
 | |
1WC9
 
 | | The crystal structure of truncated mouse bet3p | | Descriptor: | GLYCEROL, MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | Authors: | Kim, Y.-G, Lee, H.-S, Sacher, M, Oh, B.-H. | | Deposit date: | 2004-11-10 | | Release date: | 2004-12-13 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
199D
 
 | | Solution structure of the monoalkylated mitomycin c-DNA complex | | Descriptor: | CARBAMIC ACID 2,6-DIAMINO-5-METHYL-4,7-DIOXO-2,3,4,7-TETRAHYDRO-1H-3A-AZA-CYCLOPENTA[A]INDEN-8-YLMETHYL ESTER, DNA (5'-D(*(DI)P*CP*AP*CP*GP*TP*CP*(DI)P*T)-3'), DNA (5'-D(*AP*CP*GP*AP*CP*GP*TP*GP*C)-3') | | Authors: | Sastry, M, Fiala, R, Lipman, R, Tomasz, M, Patel, D.J. | | Deposit date: | 1994-12-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monoalkylated mitomycin C-DNA complex.
J.Mol.Biol., 247, 1995
|
|
1J0J
 
 | |
1BDM
 
 | |
