8GAD
 
 | |
6JCC
 
 | | structure of a de novo protein D_1CY5_M1 | | Descriptor: | Computational designed protein based on evolution | | Authors: | Meng, W, Feng, T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | structure of a computationally designed mutant protein D_1CY5_M1
To Be Published
|
|
8F6Q
 
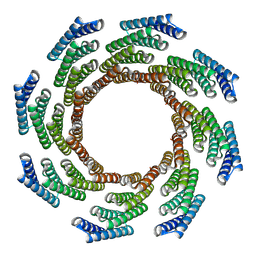 | | CryoEM structure of designed modular protein oligomer C8-71 | | Descriptor: | C8-71 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
3LTB
 
 | | X-ray structure of a non-biological ATP binding protein determined in the presence of 10 mM ATP at 2.6 A after 3 weeks of incubation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LT8
 
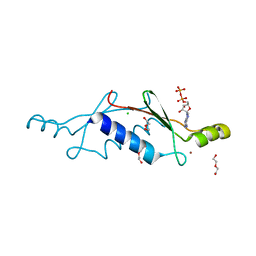 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding, crystallized in the presence of 100 mM ATP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LT9
 
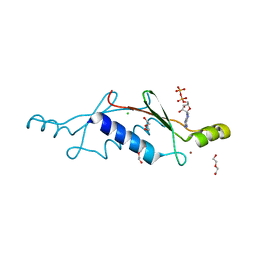 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
4KYB
 
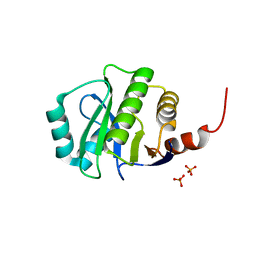 | | Crystal Structure of de novo designed serine hydrolase OSH55.14_E3, Northeast Structural Genomics Consortium Target OR342 | | Descriptor: | Designed Protein OR342, PHOSPHATE ION | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Raja, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR342
To be Published
|
|
1DOF
 
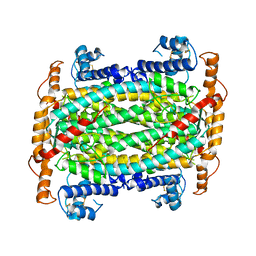 | | THE CRYSTAL STRUCTURE OF ADENYLOSUCCINATE LYASE FROM PYROBACULUM AEROPHILUM: INSIGHTS INTO THERMAL STABILITY AND HUMAN PATHOLOGY | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T.O, Goedken, E, Dixon, J.E, Marqusee, S. | | Deposit date: | 1999-12-20 | | Release date: | 2001-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
1EIX
 
 | | STRUCTURE OF OROTIDINE 5'-MONOPHOSPHATE DECARBOXYLASE FROM E. COLI, CO-CRYSTALLISED WITH THE INHIBITOR BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, OROTIDINE 5'-MONOPHOSPHATE DECARBOXYLASE | | Authors: | Harris, P, Poulsen, J.C.N, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-02-29 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the catalytic mechanism of a proficient enzyme: orotidine 5'-monophosphate decarboxylase.
Biochemistry, 39, 2000
|
|
1F1O
 
 | | STRUCTURAL STUDIES OF ADENYLOSUCCINATE LYASES | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T. | | Deposit date: | 2000-05-19 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
9BNH
 
 | |
9BNI
 
 | |
1UUO
 
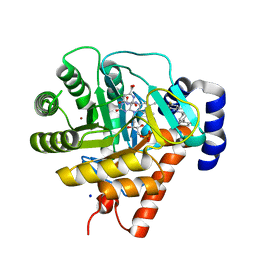 | | Rat dihydroorotate dehydrogenase (DHOD)in complex with brequinar | | Descriptor: | 6-FLUORO-2-(2'-FLUORO-1,1'-BIPHENYL-4-YL)-3-METHYLQUINOLINE-4-CARBOXYLIC ACID, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hansen, M, Le Nours, J, Johansson, E, Antal, T, Ullrich, A, Loffler, M, Larsen, S. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Inhibitor Binding in a Class 2 Dihydroorotate Dehydrogenase Causes Variations in the Membrane-Associated N-Terminal Domain
Protein Sci., 13, 2004
|
|
1UW1
 
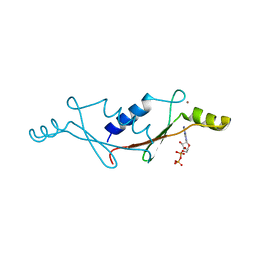 | |
1UUM
 
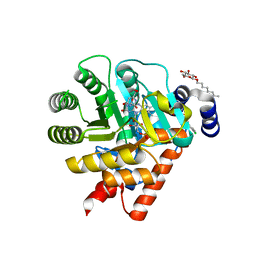 | | Rat dihydroorotate dehydrogenase (DHOD)in complex with atovaquone | | Descriptor: | 2-[4-(4-CHLOROPHENYL)CYCLOHEXYLIDENE]-3,4-DIHYDROXY-1(2H)-NAPHTHALENONE, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hansen, M, Le Nours, J, Johansson, E, Antal, T, Ullrich, A, Loffler, M, Larsen, S. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor Binding in a Class 2 Dihydroorotate Dehydrogenase Causes Variations in the Membrane-Associated N-Terminal Domain
Protein Sci., 13, 2004
|
|
8BCT
 
 | | X-ray crystal structure of a de novo selected helix-loop-helix heterodimer in a syn arrangement, 26alpha/26beta | | Descriptor: | 26alpha, 26beta, ACETATE ION, ... | | Authors: | Naudin, E.A, Mylemans, B, Smith, A.J, Savery, N.J, Woolfson, D.N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology.
Acs Synth Biol, 12, 2023
|
|
1GSO
 
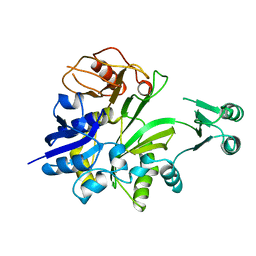 | |
1M3W
 
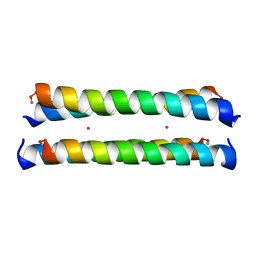 | | Crystal Structure of a Molecular Maquette Scaffold | | Descriptor: | H10H24, MERCURY (II) ION | | Authors: | Huang, S.S, Gibney, B.R, Stayrook, S.E, Dutton, P.L, Lewis, M. | | Deposit date: | 2002-07-01 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Structure of a Maquette Scaffold
J.Mol.Biol., 326, 2003
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0C
 
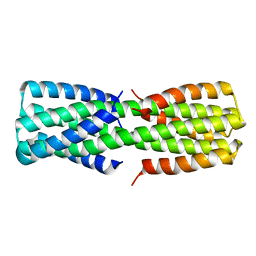 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8C3W
 
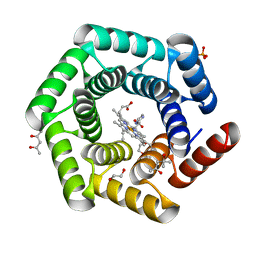 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
1K4K
 
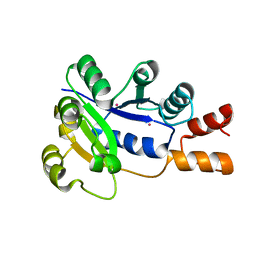 | | Crystal structure of E. coli Nicotinic acid mononucleotide adenylyltransferase | | Descriptor: | Nicotinic acid mononucleotide adenylyltransferase, XENON | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A.L. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
1KNP
 
 | |
2BXV
 
 | | Dual binding mode of a novel series of DHODH inhibitors | | Descriptor: | 2-({[3-FLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENT-1-ENE-1-CARBOXYLIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Baumgartner, R, Walloschek, M, Karlik, M, Gotschlich, A, Tasler, S, Mies, J, Leban, J. | | Deposit date: | 2005-07-27 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J. Med. Chem., 49, 2006
|
|
4B72
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (6R)-6-(4-methoxyphenyl)-2-methyl-6-(3-pyrimidin-5-ylphenyl)pyrrolo[3,4-d][1,3]thiazol-4-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
