8CUM
 
 | | X-ray crystal structure of OXA-24/40 in complex with sulfonamidoboronic acid 6d | | Descriptor: | 3-({[(1S)-1-boronopropyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, SULFATE ION | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CV7
 
 | | Peptide 2.2E in complex with BRD2-BD2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, Isoform 3 of Bromodomain-containing protein 2, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV4
 
 | | Peptide 4.2C in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
5JS5
 
 | | Nitric oxide complex of the L16F mutant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
6GC5
 
 | |
6NL3
 
 | | Solution structure of human Coa6 | | Descriptor: | Cytochrome c oxidase assembly factor 6 homolog | | Authors: | Naik, M.T, Soma, S, Gohil, V. | | Deposit date: | 2019-01-07 | | Release date: | 2019-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase.
Cell Rep, 29, 2019
|
|
7QCU
 
 | | Structure of the MUCIN-2 Cterminal domains partially deglycosylated. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mucin-2, ... | | Authors: | Gallego, P, Hansson, G.C. | | Deposit date: | 2021-11-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins.
Nat Commun, 14, 2023
|
|
7QCN
 
 | |
7QCL
 
 | | Structure of the MUCIN-2 Cterminal domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mucin-2, ... | | Authors: | Gallego, P, Hansson, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins.
Nat Commun, 14, 2023
|
|
5KLI
 
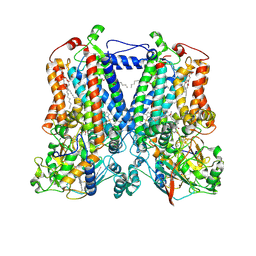 | | Rhodobacter sphaeroides bc1 with stigmatellin and antimycin | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
7MLE
 
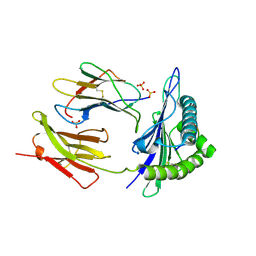 | | Crystal Structure of HLA-A*03:01 in complex with VVRPSVASK, an 9-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologous peptides derived from influenza A, B and C viruses induce variable CD8 + T cell responses with cross-reactive potential.
Clin Transl Immunology, 11, 2022
|
|
6KKO
 
 | |
8DNQ
 
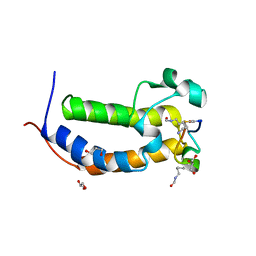 | | BRD2-BD1 in complex with cyclic peptide 2.2B | | Descriptor: | Bromodomain-containing protein 2, Cyclic peptide 2.2B, GLYCEROL | | Authors: | Patel, K, Franck, C, Mackay, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
5KKZ
 
 | | Rhodobacter sphaeroides bc1 with famoxadone | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, ASCORBIC ACID, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
6Y33
 
 | | Streptavidin mutant S112R with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y2M
 
 | | Streptavidin mutant S112R with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6TM5
 
 | |
6Y2T
 
 | | Streptavidin wildtype with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y3Q
 
 | | Streptavidin mutant S112R_K121E with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | SULFATE ION, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y25
 
 | | Streptavidin mutant S112R,K121E with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y34
 
 | | Streptavidin wildtype with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
8QMZ
 
 | | Soluble epoxide hydrolase in complex with RK4 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-2-(4-methylphenyl)sulfonyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-5-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
6IQQ
 
 | |
7L0O
 
 | |
1CCR
 
 | | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Ochi, H, Hata, Y, Tanaka, N, Kakudo, M, Sakurai, T, Aihara, S, Morita, Y. | | Deposit date: | 1983-03-14 | | Release date: | 1983-04-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of rice ferricytochrome c at 2.0 A resolution.
J.Mol.Biol., 166, 1983
|
|
