1MNU
 
 | | UNLIGANDED BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS | | Descriptor: | CADMIUM ION, PROTEIN (IGG2A-KAPPA ANTIBODY MN12H2 (HEAVY CHAIN)), PROTEIN (IGG2A-KAPPA ANTIBODY MN12H2 (LIGHT CHAIN)) | | Authors: | Van Den Elsen, J, Vandeputte-Rutten, L, Kroon, J, Gros, P. | | Deposit date: | 1999-04-30 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bactericidal antibody recognition of meningococcal PorA by induced fit. Comparison of liganded and unliganded Fab structures.
J.Biol.Chem., 274, 1999
|
|
1MCD
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-D-PHE-B-ALA-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCF
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1NCW
 
 | |
1NGP
 
 | |
1OTS
 
 | | Structure of the Escherichia coli ClC Chloride channel and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (heavy chain), Fab fragment (light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-22 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1P69
 
 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (P417S MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1PG7
 
 | | Murine 6A6 Fab in complex with humanized anti-Tissue Factor D3H44 Fab | | Descriptor: | humanized antibody D3H44, murine antibody 6A6 Fab fragment | | Authors: | Eigenbrot, C, Meng, Y.G, Krishnamurthy, R, Lipari, M.T, Presta, L, Devaux, B, Wong, T, Moran, P, Bullens, S, Kirchhofer, D. | | Deposit date: | 2003-05-27 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into how an anti-idiotypic antibody against D3H44 (anti-tissue factor antibody) restores normal coagulation.
J.Mol.Biol., 331, 2003
|
|
1NBV
 
 | |
1NAM
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
1OB1
 
 | | Crystal structure of a Fab complex whith Plasmodium falciparum MSP1-19 | | Descriptor: | ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pizarro, J.C, Chitarra, V, Verger, D, Holm, I, Petres, S, Dartville, S, Nato, F, Longacre, S, Bentley, G.A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Fab Complex Formed with Pfmsp1-19, the C-Terminal Fragment of Merozoite Surface Protein 1 from Plasmodium Falciparum: A Malaria Vaccine Candidate
J.Mol.Biol., 328, 2003
|
|
1NGQ
 
 | | N1G9 (IGG1-LAMBDA) FAB FRAGMENT | | Descriptor: | N1G9 (IGG1-LAMBDA), SULFATE ION | | Authors: | Mizutani, R, Satow, Y. | | Deposit date: | 1995-06-23 | | Release date: | 1996-07-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structures of the Fab fragment of murine N1G9 antibody from the primary immune response and of its complex with (4-hydroxy-3-nitrophenyl)acetate.
J.Mol.Biol., 254, 1995
|
|
1ND0
 
 | |
1NBZ
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K97A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NEZ
 
 | | The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, Y, Xiong, Y, Naidenko, O.V, Liu, J.H, Zhang, R, Joachimiak, A, Kronenberg, M, Cheroutre, H, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2002-12-12 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Immunity, 18, 2003
|
|
1NN8
 
 | | CryoEM structure of poliovirus receptor bound to poliovirus | | Descriptor: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | Authors: | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
1OTU
 
 | | Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1P7K
 
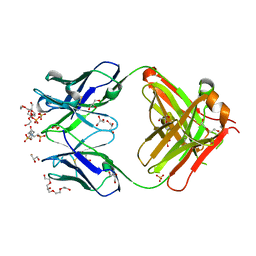 | | Crystal structure of an anti-ssDNA antigen-binding fragment (Fab) bound to 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuermann, J.P, Henzl, M.T, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2003-05-02 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an anti-DNA fab complexed with a non-DNA ligand provides insights into cross-reactivity and molecular mimicry.
Proteins, 57, 2004
|
|
1P6A
 
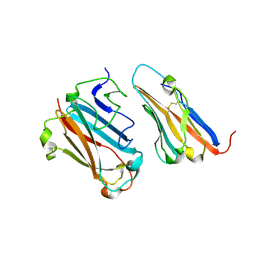 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1N0X
 
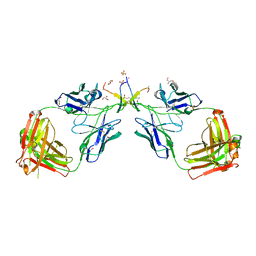 | | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, B2.1 peptide, GLYCEROL, ... | | Authors: | Saphire, E.O, Montero, M, Menendez, A, Irving, M.B, Zwick, M.B, Parren, P.W.H.I, Burton, D.R, Scott, J.K, Wilson, I.A. | | Deposit date: | 2002-10-15 | | Release date: | 2004-04-13 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope
To be Published
|
|
1N6Q
 
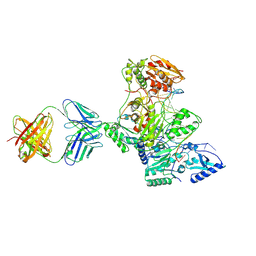 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
1PLG
 
 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
1OTT
 
 | | Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1Q9O
 
 | | S45-18 Fab Unliganded | | Descriptor: | MAGNESIUM ION, S45-2 Fab (IgG1k) heavy chain, S45-2 Fab (IgG1k) light chain | | Authors: | Nguyen, H.P, Seto, N.O, MacKenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2003-08-25 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
1QNZ
 
 | | NMR structure of the 0.5b anti-HIV antibody complex with the gp120 V3 peptide | | Descriptor: | 0.5B ANTIBODY (HEAVY CHAIN), 0.5B ANTIBODY (LIGHT CHAIN), GP120 | | Authors: | Tugarinov, V, Zvi, A, Levy, R, Hayek, Y, Matsushita, S, Anglister, J. | | Deposit date: | 1999-10-26 | | Release date: | 2000-06-03 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Anti-Gp120 Antibody Complex with a V3 Peptide Reveals a Surface Important for Co-Receptor Binding
Structure, 8, 2000
|
|
